International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013
ISSN 2229-5518
709
Yusra A. Sroor, Loay E. George, Haider G. Hussein
Keywords—Pap smear, cervical region, image processing, microscope image, biomedical image
—————————— ——————————
The automated detection and segmentation of cell nuclei in Pap smear images is one of the most interesting fields in cyto- logical image analysis as observed by Plissiti et. al.
[1]. There is a high degree of cell overlap in such images, the
presence of more than one nucleus in a cell and the lack of ho- mogeneity in image intensity. In addition, the nucleus is a very important structure within the cell and it presents significant changes when the cell is affected by a disease and thus the accu- rate definition of the nucleus boundary is a crucial task. The identification and quantification of these changes in the nucleus morphology and density contribute in the discrimination of nor- mal and abnormal cells in Pap smear images.
The segmentation of nuclei in cytological images has been studied by several researchers [2-8].
In this paper we present a method for analysis of Pap smear images based on Segmentation and ROIs extraction. The nucleus area and its surrounding plasma area will be the main target of the extraction process. This task is the hardest one due to many conflicting factors with the basic main requirements for the suc- cessful segmentation (like, cells overlapping, cytoplasm high variance due to Pap smear production sample.
The segmentation task is the major and the most difficult task in the Pap smear images analysis. Many analysis challenges are stimulated due to characteristics may appear in the micro- scopic Pap smear images, The color variations in different cells regions, this due to in-accurate staining process, The occurrence
————————————————
*Yusra A. Sroor has Master's Degree in Computer Science/University of Baghdad- Iraq. Email ysra.sroor@yahoo.com
*Loay E. George has PH.D Degree in Computer Science/University of Baghdad-
Iraq. Email loayedwar57@yahoo.com
*Haider G. Husse, FICMS(Pathology), received a fellowship degree in the field of pathology from the Iraqi Council of Medical Specialties and a membership at the international academy of cytopathology/ Central Public Health Laboratory/
Ministry of Health. Email hayderghazi@live.com
of cells overlapping areas, The appearance of false regions (i.e., as dark areas) at some of the boundary regions of the cytoplasm areas.
The images used in proposed system are Pap smear prepared and processed at the Pathology Division/ Department of the Central Public Health Laboratory in Baghdad by a newer
method (Liquid Based Cytology) (LBC) technique, where the
cells are transferred to a preservative solution prior to slide prepa- ration, which yields specimen with less cell overlap, and consid- erably enhances slide quality, stained cervical cell images for further analysis, acquired through a canon power-shot G5 digital camera adapted to an Olympus optical microscope/model Bx41. The view is magnificated by 400 (high power field) and images were stored in bitmap format and having a size 2000×1600 pixels.
The segmentation and extraction process was designed to
consist of the following steps:
As a first step toward making segmentation for the nucleus area (i.e., the ROI) and the surrounding cytoplasm, the segmenta- tion was conducted using modified k-means clustering algorithm. After analyzing large number of pap smear microscope images it was noticed that there are at least 4 types of regions, the most darkest two regions represent parts of the nucleus areas. Since the cytoplasm areas appear with bright color, so the third type of re- gions was set bright. The white color was chosen as the type be- cause it represents the non-cellular area.
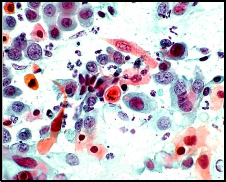
As shown in figure1, since two hue values (i.e., blue and red) may appear as the colors of the nucleus and cytoplasm areas, so the clustering algorithm was applied such that the initialized centroid values {CnB(), CnG(), CnR()} were taken such that they lead to gray colors (i.e., deep dark, dark, bright, and white), and the determination of the new centroid values, during the each round of the k-mean algorithm, is constrained to be not highly deflected from the old values and not be far from the gray.
IJSER © 2013
IJSER © 2013
http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013
ISSN 2229-5518
710

This constraint is applied by making selective distribute for the image pixels, where only the pixels that are close, up to some extent, to the nearest centroid are taken into consideration for determining the new set of centroids values, this selective distribute is controlled by two threshold values (i.e., Thr1 & Thr2); the first one is for the allowed deflection between the col- or components of the tested pixel with the nearest centroid value, while the second threshold is for bounding the allowed deflection of the gray deflection value. Due to the imposed constraints not all the image pixels are distributed among the clusters; only those close to the nearest centroid are nominated as members of the new clusters. Also, the two bounding thresholds values are re- duced from one round to the next one in order to avoid the occur- rence of large incremental drifts in the colors of centroid value.
As a last step in the introduced clustering method, a non- constrained clustering round is applied, because all the image
pixels should be indexed as members of one of the clusters. The color quantization based on constrained k-means algorithm.
Also in this stage, the color component values of the boundary value separating the non-cellular region from the cellu-
lar one is assessed using the following:
Cbckgrnd=C(NoCluster-2)+0.6{C(NoCluster-1)- C(NoCluster-2)} ,…( 1)
Where C() represents the color components of the cen- troids (i.e., CnB(), CnG, CnR()}
Figure2 presents a sample of the color quantization results;
the number of clusters was taken 4.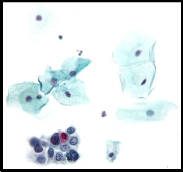
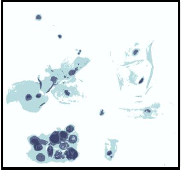
In this stage, the nominated region of interest areas (i.e., the nucleolus areas) should be flagged from other areas. Since the nucleolus areas (especially the cancerous areas) appear always dark areas so only the pixels whose gray (or intensity) value is below certain predefined threshold value (i.e., MaxBrightness) are nominated as ROI members. Also, in this stage the boundary color values that separating the white background color (i.e., the non-cellular region color) from the colors of other regions was determined using the following equations:
Cnon-nucleus = C(M) + 0.6{C(M+1) - C(M)} , ….…(2)
Where C() represents the color components of the cen- troids (i.e., CnB(), CnG, CnR()}, M is the cluster index value whose members (pixels) indexed as nucleus pixels.
Figure3 presents the output of the binarization stage that ap- plied on the segmented image shown in figure2.
This stage aims to extract the areas nominated as nucleus regions. Due to existence of overlapped areas and false areas (due to pad preparation steps) this stage was developed to make differ- ent manipulation operations in order to get as solid as possible
IJSER © 2013
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013
ISSN 2229-5518
711
nucleolus regions that are empty from gaps with possible mini- mum overlapping.
The input data to this stage is the color values of the input pap smear image {i.e., Blu(), Grn(), Red()}, the determined bina- ry index map array {i.e., Idx()} steps, and the determined non- nucleus threshold value(i.e., Cnon-nucleus). The involved steps of the nucleolus extraction process are the following:
A. Scan the index map image using seed filling algorithm to collect the aggregation on connected nucleolus pixels. The
outputs of this step are the arrays {BufX(0..Jf) & BufY(0..Jf)} which hold the column and row number of the collected pixel position, while (Jf+1) represents the number of collected points.
B. For each collected region purify it by removing the collect- ed pixels whose brightness is too high (i.e., Blue, Green, Red) > C_NonNucleus).
C. Check the size of the collected aggregation of purified pix- els: if the size is small (L<MinSize; where L is the number of pixels of the collected pixels, MinSize is a predefined pa- rameter to specify the lowest possible cell size).
D. In case the size of collected aggregation is not too small, then, determine the two dominant colors in the purified ag- gregation. This step is done by applying the following tasks: (1) Collect the mean and standard deviation of the color
components of the aggregation pixels.
(2) Determine the initial values of the expected eight cen- troid values using the mean and standard deviation values of the color components, such that:
CBlue (k) = MBlue ± σBlue
CGreen(k) = MGreen ± σGreen ,…………….(3)
CRed(k) = MRed ± σRed
Where M denotes the mean value and σ denotes the standard deviation value. The initial centroid values are either the sum or subtraction of standard deviation from the mean value, according to the above equa- tions there is eight probable combinations (i.e., k=0,1,…,7).
(3) Distribute the aggregation pixels among the eight clus- ters using the city block similarity distance measure.
(4) Choose the most populated two clusters as the initial color values for the most two dominant colors.
(5) Apply k-means algorithm to determine the best centroid values corresponding to the two dominant colors. The stopping conditions for the k-mean iterations are: (i)
when reach the predefined maximum number of itera-
tions, (ii) the differences between the new and old cen- troid values of all color components are small.
(6) Recalculate the new centroid (i.e., mean values) and the clusters dispersion (i.e., standard deviation).
(7) Determine the bounding color values for each centroid using the following:
Bmin(k)=MBlue(k) –3 σBlue , Bmax(k)= MBlue(k) + 3 σBlue
Gmin(k)=MGreen(k)–3 σGreen , Gmax(k) = MGreen(k)+ 3
σGreen ,………..(4)
Rmin(k) = MRed(k) – 3 σRed , Rmax(k) = MRed(k)+ 3 σRed E. Re-evaluate the membership of the pixels belong to the col- lected ROI. If the (R,G,B) color components of the pixel don't lay within any of the bounded range of the two domi- nants colors then the pixel is removed from the collected
ROI.
F. Filling the gaps found inside the collected ROI: in this step the collected segment will convey some gaps inside the nu- cleus body; as shown in figure (4a). To remove these artifi- cial gaps, the collected ROI is temporarily, established as a white isolated object, then, the algorithm of seed filling is applied to fill the large black hole(s) may exist within the
isolated region with white color, as shown in figure (4b).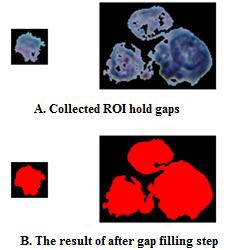
Since the seed filling algorithm has high computational complexity and to avoid its application on the small gaps, the very small gaps (i.e., consist of few pixels) are handled using tilling mechanism instead of seed filling.
This stage is auxiliary, it is applied to more accurately col- lect the pixels belong to the extracted ROI but still not considered as members of it. Till this step the thresholding mechanism was adopted decide the region membership. Since the assumption of using two dominant colors to decide the membership of all pixels lay within the area of ROI convey a sort of crisp type of deci- sions; which may lead to pixels loss especially at the boundaries of ROI because at these place the shading effect play a significant role.
In this stage additional scanning is implemented to per- form the following steps:
1. Check if the pixel doesn’t belong to ROI or not, if check re- sult is yes leave the pixel and jump to go to the next one.
While if check result is yes then continue to apply the fol-
lowing steps.
2. Check whether the ROI pixel is at the boundary of the re- gion or not. This check is done by counting the number of
IJSER © 2013
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013
ISSN 2229-5518
712
ROI pixels surrounding it. If the number of counted pixels is less more than 6 then the tested pixel is left and the scan goes to test the next pixel. Otherwise the system goes to the next steps.
3. Open a window (3x3) around the tested pixel and count the local mean and standard deviation for each of the 3 color components (Blue, Green, Red). The application of bound- ing condition of the standard deviation values, such that they should not be too small or too large.
4. Within the window's region test each pixel that flagged as not part of the pre-collected ROI using the following gradi- ent criteria:
If |Pixel.Blue-MBlue|/σBlue<R & |Pixel.Green- MGreen|/σGreen<R & |Pixel.Red - MRed|/σRed Then "The Pixel is considered as part of ROI"
Where M represents the mean value, σ represents the stand- ard deviation, and R represent the gradient value (in this work, its value is set 1.25).
5. Apply above step for certain number of times under the stopping condition "the iteration is stopped when no sur-
round non-ROI pixels are changed to be part of ROI, or the
number of iteration reach a pre- defined number"
This stage is, also, auxiliary, it is applied to remove some small artificial pores or gaps may due to gradient connectivity step. So, in this stage the segments of connected pixels (whether its index values is 0 or 1) are counted, if count values is small then the collected pixel are removed.
This stage is the final one; it is designed to do the following tasks:
1. Apply seed filling upon the index map array to collect
the flagged pixels as ROI points. These collected points are part of the nucleus area.
2. Determine the horizontal and vertical extents of the col- lected area, then, establish a sub image area to establish the ROI using the collected pixels' coordinates.
3. Scan the sub-image to allocate the border (boundary) pixels; then for each found border pixel make a local scan around it, if any pixel not flagged as nucleus pixel and its intensity below the background threshold (BckCBlu, BckCGrn, BckCRed) then pixel is consid- ered as asurrounding cytoplasm pixel.
The use of a combination consist of seed filling scanning followed by raster scanning is aimed to make fast extraction of the nucleus area with the surrounding cytoplasm.
The test results indicated that the ROI extraction process
is nearly between 86-88%, the failure is mainly occurred in two ways:
1. The appearance of small internal or as a marginal gaps. This artifact will reduce the discrimination power of some geo-
metrical features in comparison with textural features,
2. Some non-cellular dark areas are considered as ROIs areas.
To handle this problem an additional class called "Artificial ROI" was added to the list of ROI classes. And, depending on the utilization of some local texture features (like, con- currence or roughness) this class could be signified from other classes due to the existence of high local correlation between the adjacent pixels".
[1] Plissiti M.E., Charchanti A., Krikoni O. and Fotiadis D.I., “Automated segmentation of cell nuclei in PAP smear im- ages”, ITAB Proceedings International Special Topic Conference on Information Technology in Biomedicine, Greece, Ionnia, 26-28, October 2006.
[2] Bamford P., Lovell B., “Unsupervised cell nucleus segmen- tation with active contours”, Signal Processing 71(2), pp.
203-213, 1998.
[3] Bamford P., Lovell B., “A water immersion algorithm for cytological image segmentation”, Proceedings of the APRS Image segmantation workshop, pp. 75-79, University of Technology Sydney, Sydney 1996.
[4] Mouroutis T., Roberts S. J., “Robust cell nucleisegmentation using statistical modelling”, IOP Bioimaging, 6, pp. 79-91,
1998.
[5] Garrido A., Perez de la Blanca N., “Applying deformable
templates for cell image segmentation”, Pattern Recognition
33, pp. 821-832, 2000.
[6] Lee K.M., Street W.N., “Learning shapes for automatic im- age segmentation”, Proc. INFORMS-KORMS Conference, pp. 1461-1468, Seoul, Korea, June 2000.
[7] Begelman G., Gur E., Rivlin E., Rudzsky M., Zalevsky Z., “Cell nuclei segmentation using fuzzy logic engine”, Inter- national Conference on Image Processing, Vol. 5, pp 2937-
2940, October 2004.
[8] M. Lipi, N. Dilip and N. Chandan, "Cervix Cancer Diagno- sis from Pap Smear Images Using Structure Based Segmen- tation and Shape Analysis", Journal of Emerging Trends in Computing and Information Sciences, Vol. 3, No. 2, Febru- ary 2012.
IJSER © 2013
IJSER © 2013 http://www.ijser.org