International Journal of Scientific & Engineering Research, Volume 5, Issue 6, June-2014 1108
ISSN 2229-5518
Investigating UBM Degradation Products as a Possible Therapeutic Treatment for Regenerative Medicine Using a P19 Cell Model
George Iwaoka, Daniel Radin, Donna Leonardi
Abstract— Urinary Bladder Matrix (UBM) is the decellularized product of the extracellular matrix of a pig bladder and it has been shown in prior studies to promote soft tissue regeneration. However, UBM’s mechanism of action is poorly understood. P19 cells were introduced as a model cell line to elucidate these events in vitro. Three cell cultures were established from the pluripotent P19 cells: undifferentiated P19 cells, UBM-differentiated P19 cells, and spontaneously-differentiated P19 cells, referred as P19SSEA-1, P19UBM, and P19GFAP respectively. Novel results from this experiment demonstrated that UBM induces differentiation in P19 cells. These cells expressed markers for MAP2, O1, and GFAP while spontaneously differentiated cells were only positive for GFAP. Proliferation and migration assays were conducted investigating the two aspects of tissue reconstruction against UBM. All three cell cultures had increased viability across the concentrations of exogenous UBM (p<0.05) except P19UBM cells which demonstrated reduced viability at lower concentrations (25,
50µg/mL) (p<0.05). Migration results against UBM showed an increase by up to 2053% in P19SSEA-1 cells (p<0.05) and a 414% increase in P19GFAP cells (p<0.05) while P19UBM cells demonstrated reduced chemotaxis (p<0.05). The project also found evidence that each cell type utilizes different signaling pathways for chemotaxis and proliferation though the details are yet to be understood.
Index Terms— Urinary Bladder Matrix, ECM, MMP, TIMP, Embryonal Carcinoma, SSEA-1, GFAP, Pepsin, Regenerative Medicine
1 INTRODUCTION
—————————— ——————————
Recently, researchers at the McGowan’s Institute for Regenerative Medicine at the University of Pittsburgh discov- ered a substance that has potential as a novel treatment in the field of regenerative medicine. Known as Urinary Bladder Matrix (UBM), this substance is defined as the extracellular matrix (ECM) of porcine bladder [5], [9], [18]. Previous in vitro research has demonstrated that UBM degradation products are capable of causing chemotaxis and proliferation of stem cells, while demonstrating significantly reduced chemotaxis in terminally differentiated cells such as epithelial cells [18]. Ear- ly clinical trials have even shown that degradation products of UBM are capable of supporting and accelerating regeneration of soft tissues in humans. Other studies have shown that UBM degradation products have angiogenic, antimicrobial, and chemoattractive properties that are promising for future ther- apeutic use as an administrative drug [5], [18], [19]. Research- ers at the University of Pittsburgh have used the matrix to treat mouse phalanx amputation resulting in the accumulation of multipotent cells capable of neuroectodermal as well as mesodermal differentiation, which are also capable of recon- structing the entire phalanx. Additionally in 2010, they report- ed that they had successfully reconstructed the inner layer of the esophagus in 5 patients, after resection of the esophageal mucosa and submucosa, utilizing the matrix to treat patients with Barrett's esophagus and esophageal adenocarcinoma [3]. Scientists are currently researching whether they can manipu- late degraded ECM products to allow not just soft tissue heal- ing, but regeneration on a larger scale such as reconstructing entire body parts or organ systems that may be damaged or infected with a disease such as cancer. At this time however, the mechanism of action of UBM with regard to its regenera- tive potential is still not entirely understood [2], [17], [26]. This is a new and exciting concept in the field of stem cell biology,
and because the components of the matrix and their mecha- nisms are still poorly understood, the goal of this project was to investigate how the UBM degradation products may aid regeneration at a macromolecular level.
————————————————
• George Iwaoka is currently pursuing a dual undergraduate degree in Cell and Developmental Biology and Finance at the University of Rochester. E-mail: giwaoka@u.rochester.edu
• Daniel Radin is currently pursuing an undergraduate degree in Biochemistry at the University of Rochester. E-mail: danradin1@gmail.com
• Donna Leonardi, MS is currently the Director of the Bergen County Acade- mies Laboratoru of Cell Biology. E-mail:donleo@bergen.org
The extracellular matrix is a network of collagens, proteoglycans, and glycosaminoglycans that allow structural support for multicellular structures such as organs, while it regulates and maintains growth factors [22]. In vivo, degrada- tion products of ECM are crucial in the process of wound heal- ing where the proteins matrix metalloproteinases (MMPs) and protease inhibitors called tissue inhibitors of metalloprotein- ases (TIMPs) are responsible for the release and modulation of growth factors derived from degraded ECM. This modulation contributes to cell proliferation, migration, and differentiation which are all part of wound healing [13]. During an injury, the body reacts through eliciting inflammation to control infec- tion. Inflammatory cells along with macrophages and stromal cells from the edge of the wounded tissue are known to ex- press increased amounts of MMPs, which cleave parts of ECM such as collagen or elastin, causing the release of various growth factors which accelerate wound healing [10], [14], [15], [16], [20].
Based on research thus far, the widely accepted hy- pothesis by researchers with regard to the mechanism of UBM
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 6, June-2014 1109
ISSN 2229-5518
degradation products and their potential in regenerative med- icine is that these artificially degraded ECM products attract endogenous stem cells along with progenitor cells from the body, which are then induced to differentiate into the neces- sary cell types at the site of regeneration [1], [26]. Whether these matrix degradation products themselves induce differ- entiation is unknown and is one aspect of the study reported here. It is also proposed that the UBM degradation products include growth factors as well as collagen and elastin, which also modulates the regenerative process similar to what occurs in wound healing.
In an attempt to elucidate these events and under- stand the mechanism of action of the UBM, P19 cells were used as a model system along with UBM from pig (Sus scrofa domestica). P19 cells are mouse embryonic carcinoma cells (EC cells) derived from murine teratocarcinomas, or tumors in mouse embryos. These cells were used to model for stem cells as they have been used in prior studies due to their pluripo- tency [21], [23], [24]. P19 cells are capable of maintaining their pluripotency in vitro by producing self-renewal factors other than LIF [12]. P19 cells are known to differentiate in vitro into all three germ layers, including a variety of cells resembling neurons, muscle cells, and fibroblast-like cells depending on the chemical added and whether they are grown as aggregates or not [11], [12], [23], [24], [25]. Porcine bladder was chosen as a source of ECM for several reasons: its availability, cost effec- tiveness, and compatibility with humans. Pigs are highly do- mesticated and are widely available on the market. A pig’s bladder is a commercial waste product and has very minimal use and is often discarded after the pig has been euthanized when processing meat. However, most significantly, the pig exhibits similarities to humans. Pigs are mammals and their organs are of similar size. Moreover, pigs and humans share conserved areas of their major histocompatibility complexes and are often considered most appropriate for xenotransplan- tation [4]. Therefore pig bladder was seen a suitable organ for obtaining ECM degradation products.
In order to simulate degradation in vitro, pepsin was used to digest UBM to model for the MMPs degrading the ECM in vivo. Pepsin is a protease that is found in the mamma- lian stomach and is used for digestion of proteins in food into absorbable amino acids. Pepsin, rather than MMP, was used for UBM digestion because of its ability to hydrolyze a broad spectrum of proteins, including the ones hydrolyzed by MMP such as collagen and elastin [8]. Additionally, pepsin is highly cost effective and MMPs are expensive at the concentrations needed in order to digest UBM. When commercializing UBM degradation products as a treatment, an efficient, less expen- sive alternative is obviously beneficial.
Two aspects of the regenerative process were investi- gated in this project. The investigation included whether UBM degradation products induced differentiation in P19 cells, and how UBM degradation products influenced chemotaxis and proliferation in P19 cells both before and after they had been induced to differentiate. Previous studies have investigated the effects of UBM on stem cells and quiescent cells inde- pendently. However, they have never looked at the effect on a single pluripotent cell line in their differentiated and undiffer- entiated states. Nor, have studies investigated whether UBM is
alone capable of inducing differentiation in a pluripotent cell line. Therefore, three groups of cells were employed in this experiment: UBM-treated P19 cells, spontaneously differenti- ated P19 cells, and P19 cells maintained in their undifferentiat- ed state which we will refer to as P19UBM, P19GFAP, and P19SSEA-1 (Table 1), respectively (the cells were named P19GFAP and P19SSEA-1 respectively for the markers for which they were positively stained during the project—see immunocytochemistry results). The three groups of cells were then tested for chemotaxis and proliferation against UBM. The information elucidated in this project could help in the devel- opment of the artificially degraded UBM as a possible thera- peutic treatment in regenerative medicine.
Table 1
Three Cell lines Used in this Project
This table summarizes the characteristics of cells used for the duration of this experiment.
2 PROCEDURE AND METHODS
2.1 Maintenance of Cell Lines
P19 cells were obtained from American Type Culture Collec- tion (ATCC, #CRL-1825). Upon arrival of the cells, they were immediately thawed out in a 37ºC water bath for 90 seconds and transferred into a 15mL centrifuge tube. Complete media was added so the total volume was approximately 10mL. The tube was then centrifuged at 1,000 rpm for 7 minutes. A pellet formed on the bottom of the tube, and the media was aspirat- ed carefully making sure that the pellet was intact. The pellet was then resuspended into 15mL of complete media and the culture was seeded into a T-75 flask. The complete growth medium consisted of DMEM/F12 (Invitrogen #10565018),
10%FBS (Invitrogen, #10438018), and 1%Penn/Strep (Invitro- gen, #15140122). The cells were incubated at 37ºC at 5% CO2 with 100% humidity. The cells were trypsinized as needed when the flask became 80%~ confluent.
2.2 UBM Preparation/Pepsin Digest
UBM was prepared according to a protocol supplied by re- searchers at the McGowan’s Institute of Regenerative Medi- cine at the University of Pittsburgh. Fresh pig bladder was purchased from the local butcher store and was immediately stored at -20ºC upon arrival to the lab in order to lyse the cells. The bladder was then thawed out in a 37ºC water bath and the bladder was cut open and muscle layers were scraped off. The remaining product was the basement membrane and the tuni- ca propria, a rubbery semi-transparent white sheet [2], [5], [9], [17], [18]. The sheet was then washed in a 0.1% peracetic ac- id/4% ethanol solution in deionized water for 2 hours, with
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 6, June-2014 1110
ISSN 2229-5518
PBS rinses every 15 minutes for decellularizing the remaining cells on the bladder. The sheet was then snap frozen at -80ºC, lyophilized and grinded using a commercial blender to create a powder. The resulting powder was then was mixed at a ratio of 100mg of pepsin per every gram of powder in 0.01M hydro- chloric acid in order to digest components of the matrix. This resulted in a viscous gel-like substance, which was stored at -
80ºC for later use. A separate control group containing pepsin in 0.01M HCl without any UBM was created, referred to as the Pepsin Control. The Pepsin Control was used as a negative control in all the experiments.
2.3 UBM Differentiation of P19 Cells
P19 cells were investigated to see whether UBM degradation products are capable of inducing differentiation, since its iso- lated effect on cell differentiation had never been studied prior to this experiment. Pluripotent P19 cells were grown to about
80% confluency where they were then trypsinized and trans- ferred to a T-25 flask at 1x106 cells/mL. The cells were then treated with 75μg/mL of UBM in complete growth media and incubated at 37ºC at 5% CO2 and 100% humidity for 5 days, splitting and replenishing media when necessary. Pluripotent cells were also maintained in a similar fashion, but with only UBM vehicle as a control group. After the 5 day incubation period, the cells were then moved to a new sterile T-75 flask where after every 24 hours, they were given complete growth media to allow the cells to stabilize for 5 days. An immunocy- tochemistry assay was then performed to determine if the cells had differentiated. Fluorescent photos were taken under a Nikon Eclipse 80i Fluorescent Microscope (Nikon). Addition- ally, compound light microscope pictures were taken during the process to determine whether they had morphological dif- ferences at 10x magnification (pictures not shown).
2.4 Spontaneous Differentiation of P19 Cells
P19 cells naturally give off signaling molecules such as growth factors into the media inducing neighboring cells to differenti- ate if media is not replenished frequently [7]. Spontaneous differentiation was accomplished according to a published protocol in this fashion [6]. Pluripotent P19 cells were cultured with complete growth media being replenished only every 5 days and incubated at 37ºC at 5% CO2 and 100% humidity. Initially, the cells were plated in a T-75 flask at ~50% confluen- cy, and were grown as a monolayer. The cells were replated and they were allowed to stabilize for another 5 days with media being replenished minimally. An immunocytochemis- try assay was then performed to ensure differentiation of the cells. Photos were also taken under a compound light micro- scope during the process to determine whether they had dif- ferentiated morphologically (pictures not shown).
2.5 Immunocytochemistry
P19SSEA-1, P19GFAP, and P19UBM cells were tested for spe- cific markers each time an assay was performed to confirm the cell identity as either being pluripotent or differentiated. A Neural Stem Cell Characterization Kit (Millipore, SCR019) was used and the cell lines were tested for five markers: SSEA-1, MAP2, Nestin, GFAP, and O1 (Table 2). The cells were grown in 8-well chamber slides (Thermo Scientific, #154534) over-
night at 100,000 cells/chamber (5x105 cells/mL). The following day, the cells were incubated in 4% paraformaldehyde in PBS for 30-40 minutes at room temperature after aspirating the media. The cells were then rinsed three times in PBS. A block- ing solution consisting of 5% goat serum and 0.3% Triton X-
100 in 1% PBS with primary antibodies directed against each of the five markers were added and were then stored over- night at 4ºC (Mouse anti-Nestin, 5ng/μL Mouse anti- MAP2,
5ng/μL, Mouse anti-O1, 2ng/μL, Rabbit anti-GFAP). As a negative control group, equivalent concentrations of mouse IgG, rabbit IgG, or mouse IgM was used corresponding to each isotype for the markers tested. The next day, the cells were washed two times with 1X PBS and two more times with appropriate blocking solution. An Alexa Fluor secondary anti- body (Invitrogen) correspondent to each primary antibody was then added at a dilution factor of 1:500 for 2 hours at room temperature. The solution was aspirated again and rinsed in 1X PBS two more times. The 8-well plate was then fixed and washed with water after applying anti-fade. The plates were then visualized under a fluorescent microscope at either 594 (emission 617nm) or 488 nm (emission 519nm) de- pending on the secondary antibody used. The table below de- lineates the positive staining used to characterize the state of differentiation of the cells as per manufacturer’s protocol.
Table 2
Immunocytochemistry Markers
| SSEA-1 | Nestin | MAP-2 | GFAP | O1 |
Pluripotent Cell Line Neural Stem Cell Neuronal Lineage Astrocyte Lineage Oligoden- dride Cells | + + - - - | - + - - - | - - + - - | - - - + - | - - - - + |
This table demonstrates what immunocytochemistry markers correspond to which cell lineages or cell types.
2.6 Eulate Preparation
UBM eulates were prepared according to a protocol supplied to us by researchers at the McGowan’s Institute of Regenera- tive Medicine at the University of Pittsburgh. Eulates were created to determine whether the cells were reacting to the smaller water-soluble components in the degraded UBM, such as various growth factors, rather than water insoluble compo- nents such as collagen or elastin released during the enzymatic digestion of UBM. 100mg of UBM powder was placed in each well. 3mL of DMEM/F12 were then added to each of the wells and was incubated for 20-24 hours at 37ºC at 5% CO2 and
100% humidity. The solution was then collected and centri- fuged at 1200 rpm, and the powder was collected as a pellet
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 6, June-2014 1111
ISSN 2229-5518
and discarded. The supernatant was sterilized by using a sy- ringe filter and stored at -80ºC until used for eulate assays. Basal media consisting of just DMEM/F12 was used as a nega- tive control group to the eulates.
2.7 Migration Assay
A migration assay was conducted using 96-well Chemotaxis
Chambers (NeuroProbe, #MBA96). The cells were grown to
80-90% confluence and then “starved” for a day by placing
them in a starvation media consisting of DMEM/F12 with 1%
Penn/Strepp without any FBS. Cells were then trypsinized
and prepared for plating at 30,000 cells/well in the 96-well
upper chamber of the NeuroProbe device (6x105 cells/mL).
The UBM was prepared at various concentrations ranging
from 200μg/mL-500μg/mL. 400μL of each UBM concentration was placed in each of the wells in the bottom chamber of the device. The top and bottom wells were separated with an 8μm filter pore (NeuroProbe, #PFD8) that had been soaked with a
solution consisting of 0.05mg/mL of collagen I in a 0.02M ace- tic acid solution overnight. The cells were incubated for 24 hours at 37ºC and 5% CO2. After 24 hours, the filter with at- tached cells was then fixed in 95% methanol for 2 minutes, and was stained using 4% Giemsa Stain in Gurr buffer (pH of 6.8). The filter was then carefully washed with distilled water in order to wash off any excess residue from staining. The filter was then mounted onto a glass slide and cells were counted for analysis using a Nikon compound light microscope.
2.8 Data Analysis
Data were analyzed using Microsoft Excel (Microsoft). All as- says were repeated at least twice with a sample size of n=3 or more. A Student’s t-test was used to determine significance and alpha was set at 0.05. Data are represented as the mean ± standard deviation.
3 RESULTS
P19 Differentiation
Pictures from the tissue culture microscopes (100x) showed that both P19GFAP and P19UBM showed morphological dif- ferences after both cell lines had been induced to differentiate. P19GFAP cells displayed processes that resembled dendrites after they had gone through spontaneous differentiation (pic- tures not shown). On the other hand, P19UBM cells, which were observed to grow in aggregates, showed a different phe- notype with regards to both morphology and aggregate as- sembly after being exposed to UBM for 5 days. Some aggre- gates showed dendrite-like processes similar to P19GFAP cells, while other cells formed aggregate formations that were different from the ones that were seen in their undifferentiated state.
Immunocytochemistry

An immunocytochemistry assay was conducted after P19 cells had been either exposed to UBM for 5 days, had been allowed to spontaneously differentiate, or were maintained for plurip- otency. Results demonstrated that P19 cells that were allowed to spontaneously differentiate were observed to be GFAP pos-
itive, while P19 cells that remained undifferentiated were SSEA-1 positive, demonstrating that these untreated cells were pluripotent (Table 3). Interestingly, P19 cells which had been exposed to UBM were shown to be positive for the marker GFAP, MAP-2, and O1, possibly demonstrating that these cells were following multiple pathways of neuronal differentiation or a different rate of differentiation (Table 3). Both P19GFAP and P19UBM were negative for the marker SSEA-1 meaning they had lost their pluripotent property. Immunostaining was performed before all viability and migration assays for the three cell lines to ensure their cell type before each assay.
Table 3
Immunocytochemistry Results
| SSEA-1 | Nestin | MAP-2 | GFAP | O1 |
P19SSEA-1 P19GFAP P19UBM | + - - | - - - | - - + | - + + | - - + |
This table shows the results of the immunocytochemistry analysis per- formed to determine which markers P19 subtypes still contained.
Migration Assay Results
Results from the migration assay demonstrated that UBM acts as a positive chemoattractant in P19SSEA-1 and P19GFAP cells while it showed reduced chemoattractive properties against P19UBM differentiated cells (Figure 1). After 24 hours, P19SSEA-1 cells demonstrated increased migration by up to
2053% at the highest concentration of 500μg/mL UBM com- pared to the control group exposed to only complete growth media (p<0.05). P19GFAP cell migration increased by 414% (with respect to untreated control) at its most optimal concen- tration of 300μg/mL (p<0.05). Interestingly, at higher concen- trations tested there is a decrease in chemotaxis of P19GFAP cells. Between 300μg/mL and 400 μg/mL, there is a 317% drop in migration (p<0.05). One explanation may be that the high concentrations of UBM are negatively chemotactic to P19GFAP cells. This is an important aspect to consider when applying this product as a therapeutic option, since high con- centrations of UBM could cause healthy differentiated cells inside the body to exhibit reduced chemotaxis. Conversely, P19UBM showed minimal chemotaxis in all of the concentra- tions tested. Migration results with the water extractions of eulates showed that the eulates acted as a positive chemoat- tractant for P19SSEA-1 cells and P19GFAP cells (Figure 2 on the next page). In P19SSEA-1 cells, there was a 1259% increase in migration against eulates when compared to the control group consisting of no eulates after 24 hours (p<0.0005). P19GFAP cells increased in migration by 57% under the pres- ence of the eulate (p<0.05). P19UBM cells decreased in migra- tion by 6% when compared to the control group (p<0.05).
IJSER ©
http://www.ij
International Journal of Scientific & Engineering Research, Volume 5, Issue 6, June-2014 1112
ISSN 2229-5518
Fig 1. Chemotactic effect of UBM degredation products on various cell types. Concentrations of 200-500μg/mL were used. Data represented as mean ± s.d. * denotes p<0.05 ** denotes p<0.01 when compared to the when no UBM is added (n=4)
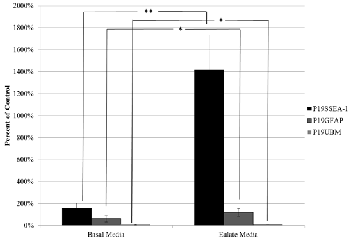
Fig 2. Effect of UBM-Eulates on the chemotaxis of various cell types.
100% migration is the average value of the cells that were grown in com- plete media. Data represented as mean ± s.d. * denotes p<0.05 ** de-
notes p<0.01 (n=5)
Viability Assay Results
Viability assays demonstrated that UBM degradation products at the concentrations tested caused increased viability in all three cell types (Figure 3) with the exception of P19UBM at the two lower concentrations. The greatest viability was seen at
75μg/mL in all concentrations, which was the closest concen- tration to the most optimal concentration of UBM reported in the literature at 78.9μg/mL [18]. These results suggest the pepsin digestion of the UBM renders molecules, which en- hance proliferation in the three cell types tested.
Viability results with eulates showed that eulates had no statistical significance with regard to viability against P19SSEA-1 cells (Figure 4). Combining this with the results in Figure 3 suggests that the molecules causing increased cell growth in the pluripotent P19SSEA-1 cells are not the small water soluble molecules present in the eulates and may be large proteins such as collagen and elastin which are only re- leased upon degradation of the UBM with pepsin. With regard
to the P19UBM cells, there was a 40% increase in viability (p<0.05) with the eulate treatment suggesting small water sol- uble products found in the eulates affected viability as they most likely did in the UBM pepsin digest. Interestingly, there was a 34% decrease in viability in the P19GFAP cells (p<0.05) treated with the eulate. Combining this with the P19GFAP results in Figure 3 suggests that the molecules causing prolif- eration of these particular differentiated cells from the pepsin digest of the UBM are most likely large and insoluble in the water extraction. Furthermore, there is actually some small water-soluble molecules in the eulate, which is blocking pro- liferation of the P19GFAP cell type. This is very interesting, having implications in therapeutic application and warrants further investigation. This leaves the possibility that different lineages of P19 cells are reacting to different components of the pepsin digested UBM as well as the eulate.
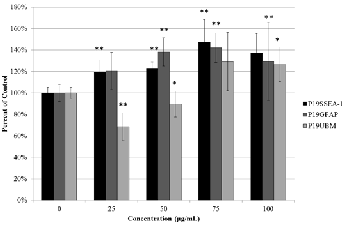
Fig 3. Effect of UBM degradation products on the viability of various cell types. Concentrations of 25-100μg/mL were used. Data represented as mean ± s.d. * denotes p<0.05 ** denotes p<0.01 when compared to the when no UBM is added. (n=5)
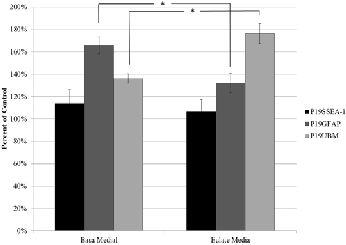
Fig 4. Effect of UBM-Eulates on viability of various cell types. 100% migra- tion is the average value of the cells that were grown in complete media. Data represented as mean ± s.d. *denotes p<0.05 **denotes p<0.01 (n=5)
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 6, June-2014 1113
ISSN 2229-5518
4 DISCUSSION
This experiment strongly suggests for the first time, that pep- sin digested UBM degradation products are capable of directly inducing differentiation in P19 cells. This novel finding sup- ports the possibility that UBM degradation products may con- tain signals that will allow stem cells to differentiate in vivo, though further tests would need to be conducted in order to confirm this hypothesis. In addition, this research supplies important data for understanding the sequence of events that occur if UBM application is employed in vivo.
In previous studies, it was believed that UBM degra- dation products acted as a positive chemoattractant and a mi- togenic agent only to undifferentiated cells. Therefore, with regards to chemotaxis, the increased migration in P19SSEA-1 and the decreased migration in P19UBM was expected. How- ever, P19GFAP cells, which had also been differentiated, had demonstrated an increased chemotactic response, contrary to hypothesized behavior based on previous reports. Moreover, a major difference between this study and previous studies is that cells used in this experiment were newly differentiated cells and their parent cells (P19SSEA-1 cells) were derived from tumors, which have abnormalities unseen in normal so- matic cells. This project, unlike previous experiments, used cells that were induced to differentiate while previous exper- iments used primary cell lines that were acquired in their dif- ferentiated state. This suggests that cells undergoing differen- tiation or shortly after they have been differentiated behave differently from primary cells with regards to their response to UBM.
Viability assays also demonstrated interesting results. It was hypothesized based on past studies that P19SSEA-1 cells would be the only cell type to significantly proliferate, and that P19GFAP and P19UBM cells would be reduced in viability, if not cytostatic, as they are both differentiated. Con- trary to the expected hypothesis, all three cell lines showed some proliferation. Furthermore, previous experiments con- ducted with epithelial cells showed that UBM products were in fact, toxic to differentiated cells at the site of the wound, therefore, decreasing proliferation and causing apoptosis nec- essary in wound remodeling. [18]. Therefore, differentiated P19 cells were hypothesized to decrease in viability or remain cytostatic. Unexpectedly, P19GFAP and P19UBM cells both proliferated in a statistically significant fashion (though P19UBM cells had slightly decreased in viability at lower con- centrations, they were not cytotoxic to the extent previously shown [18].
Data from the viability assay with the eulates also strongly suggest that the three cell types established here may be reacting to different components of the UBM. Because P19UBM cells were the only cell type to increase in viability under the presence of the eulates, these cells may be reacting to the smaller, water soluble components of the UBM, while P19GFAP cells and P19SSEA-1 cells may be reacting to larger components of the UBM that do not reside in the eulate, but do reside in the pepsin digest. Therefore, each cell type may possibly be utilizing different signaling pathways, another topic of further interest and investigation.
5 CONCLUSION
This project helped elucidate how UBM degradation products function in vitro and potentially in vivo, and strong- ly suggests evidence for a novel hypothesis on the sequence of events that may occur inside the body when UBM powder is applied to a damaged tissue or organ. Taken the results demonstrated during the experiments all together, this project hopes to provide a better understanding for the sequence of events that may occur when using pepsin digest of the porcine urinary bladder matrix with regard to its potential in regener- ative medicine. Never before have UBM products been shown to directly cause differentiation in pluripotent cells. Data from the eulate experiments also suggest that growth factors that reside in these degraded UBM products may be the key to the events that are occurring inside the body, as the eulate in- cludes only those components that are water-soluble. This excludes most of the ECM proteins such as collagen and elas- tin. Up until now, researchers have only been successful in reconstructing soft tissues both in vitro and in vivo. Therefore, it is crucial for us to understand the mechanism of action at a macromolecular level, in order to attempt larger scale recon- struction in the body, as implications for UBM as a treatment in regenerative medicine, or as a scaffold in tissue engineering are promising.
In terms of future research, investigating the mole- cules that reside within the eulates and pepsin digest would be the crucial next step in understanding how UBM degrada- tion products work. Smaller components of the UBM, mainly growth factors, are believed to be present in the eulate. By in- vestigating this solution, active growth factors could hopefully be discovered, and by then looking at which signaling path- ways are being utilized, we may start to understand UBM as an effective product. In addition, a microarray analysis of the three cell types tested in this experiment would hopefully help explain the difference between the cell types, by comparing and contrasting which genes are up regulated or down regu- lated.
Regenerative medicine is a rapidly growing field, and UBM being one of its promising products warrants further investigation in the coming years.
6 REFERENCES
[1] Agrawal V., Brown B.N., Beattie A.J., Gilbert T.W., & Badylak S.F., (2009) Evidence of Innervation following Extra- cellular Matrix Scaffold Mediated Remodeling of Muscular Tissues. J Tissue Eng Regen Med. 3(8): 590-600
[2] Badylak, S.F., (2001) Marrow-derived Cells Populate Scaf- folds Composed of Xenogeneic Extracellular matrix. ExpHe- matol. 29(11): 1310-1318.
[3] Badylak, S.F., Hoppo, T., Nieponice, A., Gilbert, T.W., Da- vison, J.M., & Jobe, B.A., (2011) Esophageal Preservation in Five Male Patients after Endoscopic Inner-Layer Circumferen- tial Resection in the Setting of Superficial Cancer: A Regenera- tive Medicine Approach with a Biological Scaffold. Tissue En-
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 6, June-2014 1114
ISSN 2229-5518
gineering Part A. (11-12): 1643-1650.
[4] Bahram S., Bresnahan, M., Geraghty, D.E., & Spies T., (1994) A second lineage of mammalian major histocompatibil- ity complex class I genes. Proc. Natl. Acad. Sci.91: 6259-6263.
[5] Brennan, E.P., Reing J., Chew, D., Myers-Irvin, J.M., Young, E.J., & Badylak, S.F., (2006) Antibacterial Activity within Deg- radation Products of Biological Scaffolds Composed of Extra- cellular Matrix. Tissue Engineering 12(10): 2949-2955
[6] Cuddenec, C.A., & Nicolas J.F., (1977) Blood formation in a clonal cell line of mouse tetracarcinoma. J. Embryol. exp. Morph., 38: 203-210
[7] Doetschman, T.C., Eistetter H., Katz, M., Schmidt, W., and Kemler, R., (1985) The in vitro development of blasteocyte- derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. J Embryol exp Morph 87:
27-45
[8] Fruton, J.S., (1970) The specificity and mechanism of pepsin action Adv Enzymol Relat Areas Mol Biol 33:401-443
[9] Gilbert, T.W., Stolz, D.B., Biancaniello, F., Simmons-Byrd, A., & Badylak, S.F. (2005). Production and characterization of ECM powder: implications for tissue engineering applications. Biomaterials, 26(12):143-1435. PMID: 15482831
[10] Gill, S.E., & Parks, W.C., (2007) Metalloproteinases and their inhibitors: regulators of wound healing. Int J Biochem Cell Biol. 40(6-7): 1334-1347
[11] Jones-Villeneuve, E.M.V., Rudnicki, M.A., Harris, J.F., & McBurney, M.W., (1983) Retinoic Acid-Induced Neural Differ- entiation of Embryonal Carcinoma Cells. Molecular and Cellu- lar Biology, 3(12) 2271-2279.
[12] Kawazoe, S., Ikeda, N., Miki, K., Shibuya, M., Morikawa, K., Nakano, S., Oshimura, M., Hisatome, I., & Shirayoshi, Y., (2009) Extrinsic Factors Derived from Mouse Embryonal Car- cinoma Cell Lines Maintain Pluripotency of Mouse Embryonic Stem Cells Through a Novel Signal Pathway. Develop, Growth Diff. 51, 81-93
[13] Matrisian, L.M., & Hogan, B.L., (1990) Growth factor- regulated proteases and extracellular matrix remodeling dur- ing mammalian development. Curr Top Dev Bio. 24:219-259
[14] Mohan, R., Chintala, S.K., Jung, J.C., Villar, W.V,L., McCabe, F., Russo, L.A., Lee, Y., McCathy, E., Wollenberg, K.R., Jester, J.V., Wang, M., Welgus, H.G., Shipley, J.M., Sen- ior, R.M., & Fini, M.E., (2002) Matrix metalloproteinase gelati- nase B (MMP-9) coordinates and effects epithelial regenera- tion. The Journal of Biological Chemistry 277(3):2065-2072.
[15] Novotna, J., & Herget, J., (2002) Possible role of matrix metalloproteinases in reconstruction of peripheral pulmonary arteries induced by hypoxia. Physiol Res 51(4):323-334
[16] Pilcher, B.K., Dumin, J.A., Sudbeck, B.D., Krane, S.M., Welgus, H.G., & Parks, W.C., (1997) The activity of colla- genase-1 is required for keratinocyte migration on a type I collagen matrix. The Journal of Cell Biology 137(6):1445-1457
[17] Przyborski, S.A., Christie, V.B., Hayman, M.W., Stewart, R., & Horrocks, G.M., (2004). Human Embryonal Carcinoma Stem Cells: Models of Embryonic Development in Humans. Stem Cells and Development, 13, 400-408
[18] Reing, J., Zhang L., Myers-Irvin, J., Cordero K., Freytes, D., Heber-Katz, E., Bedelbaeva, K., McIntosh, D., Dewilde, A., Braunhut, S., & Badylak, S., (2009). Degradation Products of Extracellular Matrix Affect Cell Migration and Proliferation. Tissue Engineering: Part A, 15(3): 604-615
[19] Sarikaya A, Record R, Wu CC, Tullius B, Badylak & S, Ladisch M., (2002) Antimicrobial activity associated with ex- tracellular matrices. Tissue Eng. 8(1):63-71. PMID: 11886655
[20] Shapiro, S.D., & Senior, R.M., (1999) Matrix metallopro- teinases matrix degradation and more. American Journal of Respiratory Cell and Molecular Biology 20:6 1100-1102
[21] Soprano, D.R., Teets, B.W., & Soprano, K.J., (2007). Role of Retinoic Acid in the Differentiation of Embryonal Carcinoma and Embryonic Stem Cells. Vitamins and Hormone, 75:69-95.
[22] Taipale, J., & Keski-Oja, J., (1997) Growth factors in the extracellular matrix. FASED J. (11) 51-59.
[23] Van der Heyden, M.A.G., & Derize, L.H.K. (2002). Twenty one years of P19 cells: what an embryonal carcinoma cell line taught us about cardiomyocyte differentiation. Cardiovascular Research, 58(2):292-302.
[24] Van der Heyden, M.A.G., Van Kempen, M.J.A., Tsuji, Y., Rook, M.B., Jongsma, H.J., & Opthof, T. (2003) P19 embryonal carcinoma cells: a suitable model system for cardiac electro- physiological differentiation at the molecular and functional level. Cardiovascular Research, 58(2): 410-422.
[25] Yoon, J.S., Lee, M.Y., Lee, J.S., Park, C.S., Youn, H, J., & Lee, J.H., (2009) Bis is Involved in Glial Differentiation of P19
Cells Induced by Retinoic Acid. Korean J Physiol Pharmacol,
13: 251-256.
[26] Zantop T, Gilbert TW, Yoder MC, & Badylak SF., (2006) Extracellular Matrix Scaffolds are Repopulated by Bone Mar- row-Derived Cells of Achilles Tendon Reconstruction. Orthop Res. 24(6):1299-309.PMID: 16649228
IJSER © 2014 http://www.ijser.org



