International Journal of Scientific & Engineering Research Volume 4, Issue 2, February-2013 1
ISSN 2229-5518
Hiding Secret Data in DNA Sequence
Debnath Bhattacharyya, Samir Kumar Bandyopadhyay
Abstract— In this paper, we propose an algorithm to hide secret message in DNA String to increase the security during transmission of data. On preparation, we consider the behavior and characteristics of DNA sequences. In this paper, we propose a new Binary Coded DNA rules towards Data Hiding in DNA. DNA String is identified randomly where a secret message is encoded at Sender’s end. Same DNA String is used at Sender’s and Receiver’s ends. At the Receiver’s end, the forwarded secret message is decoded by the help of Message Index that generates at Sender’s end.
Index Terms— Security, DNA, Data Hiding, Steganography.
—————————— ——————————
1 INTRODUCTION
TEGANOGRAPHY and watermarking are main parts of the fast developing area of information hiding. Steganog- raphy and watermarking bring a variety of very important techniques how to hide important information in an undetect-
able and/or irremovable way in audio and video data.
The main goal of steganography is to hide a message m in some audio or video (cover) data d, to obtain new data d', practically indistinguishable from d, by people, in such a way that an eavesdropper cannot detect the presence of m in d'.
Shortly, we can say that cryptography is about protecting the content of messages; steganography is about concealing its very existence.
Data Hiding is the process of secretly embedding infor- mation inside a data source without changing its perceptual quality. Data Hiding is the art and science of writing hidden messages in such a way that no one apart from the sender and intended recipient even realizes there is a hidden message. Generally, in Data Hiding, the actual information is not main- tained in its original format and thereby it is converted into an alternative equivalent multimedia file like image, video or audio which in turn is being hidden within another object. This apparent message is sent through the network to the re- cipient, where the actual message is separated from it.
The requirements of any data hiding system can be catego- rized into security, capacity and robustness Cox et al. (1996). All these factors are inversely proportional to each other creat- ing the so called data hiding dilemma. The focus of this paper aims at maximizing the first two factors of data hiding i.e. se- curity and capacity coupled with alteration detection. The proposed scheme is a data-hiding method that uses high reso- lution digital video as a cover signal. The proposed recipient need only process the required steps in order to reveal the message; otherwise the existence of the hidden information is
————————————————
Debnath Bhattacharyya is currently Professor and Head, Department of Computer Science and Engineering, FET, NSHM Knowledge Campus- Durgapur, Durgapur-713212, West Bengal, India, PH-+919903682993. E-mail: debnathb@gmail.com
Samir Kumar Bandyopadhyay is currently the Vice Chancellor, West Ben-
gal University of Technology, Salt Lake, Kolkata. E-mail: skb1@vsnl.com
virtually undetectable. The proposed scheme provides the ability to hide a significant quality of information making it different from typical data hiding mechanisms because here we consider application that require significantly larger pay- loads like video-in-video and picture-in-video.
The purpose of hiding such information depends on the application and the needs of the owner/user of the digital media. Data hiding requirements include the following:
Imperceptibility- The video with data and original data
source should be perceptually identical.
Robustness- The embedded data should survive any
processing operation the host signal goes through and
preserve its fidelity.
Capacity-Maximize data embedding payload.
Security- Security is in the key.
Data Hiding is the different concept than cryptography, but uses some of its basic principles [1].
In this paper, we have considered some important features of data hiding. Our consideration is that of embedding infor- mation into video, which could survive attacks on the net- work.
2 PREVIOUS WORKS
Mohammad Reza Abbasy, et al, in 2011, proposed [2] a data hiding method where data were efficiently encoded and de- coded following the properties of DNA sequence. Comple- mentary pair rules of DNA were used in their proposed meth- od.
Mohammad Reza Najaf Torkaman, et al, in 2011, proposed a new cryptography protocol based on DNA steganography to reduce the usage of public cryptography to exchange session key [3]. They also claimed that, the attackers were not aware of transmission of session key through unsecured channel.
Dominik Heider and Angelika Barnekow, in 2008, nicely explained the concept of DNA Watermarking [4]. They ex- plained that the DNA watermarks produced by DNACrypt do not influence the translation from mRNA into protein. By ana- lyzing the vacuolar morphology, growth rate and ability to sporulate they confirmed that the resulting Vam7 protein was functionally active.
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 4, Issue 2, February-2013 2
ISSN 2229-5518
Boris Shimanovsky, Jessica Feng, and Miodrag Potkonjak, in 2002, proposed the original idea of hiding data in DNA and RNA. They defined two techniques, firstly, hiding data in non- coding DNA and secondly, store data in active coding seg- ments [5].
3 OUR WORK
DNA sequencing is any process used to map out the sequence of the nucleotides that comprise a strand of DNA [6]. DNA nucleotides are the small repeating units that, joined together by the millions into a long spiral ladder shape, form the DNA strand, also called a DNA molecule or double helix or, simply, DNA.
A nitrogen-containing ring structure called a base. The base is attached to the 1' carbon atom of the pentose. In DNA, four different bases are found:
two purines, called adenine (A) and guanine (G)
two pyrimidines, called thymine (T) and cytosine (C)
Purines and pyrimidines are two of the building blocks of nucleic acids. Only two purines and three pyrimidines occur widely in nucleic acids. In Fig. 1, two purines and two pyrim- idines are shown only, we have not considered uracil in our coding.
Fig. 1. Structure of Purines and Pyrimidines.
TABLE 1
DNA Base Coding
Four bases in the DNA code represent a letter in our work. Abbreviations are used as given in Table 1.
A typical DNA instruction can be stated as:
STOP : Alanine + Aspartic Acid + Glycine + Alanine : STOP Build a protein by combining alanine, aspartic acid, glycine,
and alanine. It would be coded in DNA as:
TAA GCC GAT GGA GCC TAA
110000 100101 100011 101000 100101 110000
TABLE 2
DNA codons and their meanings
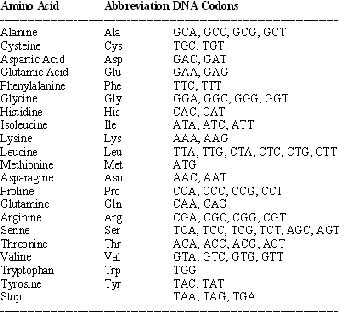
The Stop instruction is important because an organism would otherwise produce infinitely long, tangled blobs of amino acids, instead of useful proteins that perform a specific function such as transporting oxygen in blood (i.e., hemoglo- bin). DNA Codons and their corresponding amino acids are stated in Table 2.
Algorithms
Step 1: Randomly select a DNA Sequence or String from avail- able Strings. As for example, in our experiment, it is: TAA- GCCGATGGAGCCTAA
Step 2: String is DNA Coded as per our coding scheme stated in Table 1.
TAAGCCGATGGAGCCTAA =>
110000100101100011101000100101110000
Step 3: Index the DNA String.
T0A1A2G3C4C5G6A7T8G9G10A11G12C13C14TA15A16
Encoding message into DNA String
Step 1: Message, M. As for example, M is “UN64”
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 4, Issue 2, February-2013 3
ISSN 2229-5518
Step 2: Binary Coded M.
UN64 => 01010101010011100000011000000100
Step 3: DNA Coded, M, following the coding table of Table 1.
01010101010011100000011000000100 =>
CCCCCATGAACGAACA
Step 4: Message Index, CCCCCATGAACGAACA => 4444410311431141
Message Index is the first positional index value of the DNA String. For example, our DNA Coded Message is CCCCCATGAACGAACA, the first C of this message, is oc- curred at the 4th indexed position of DNA String, and so on.
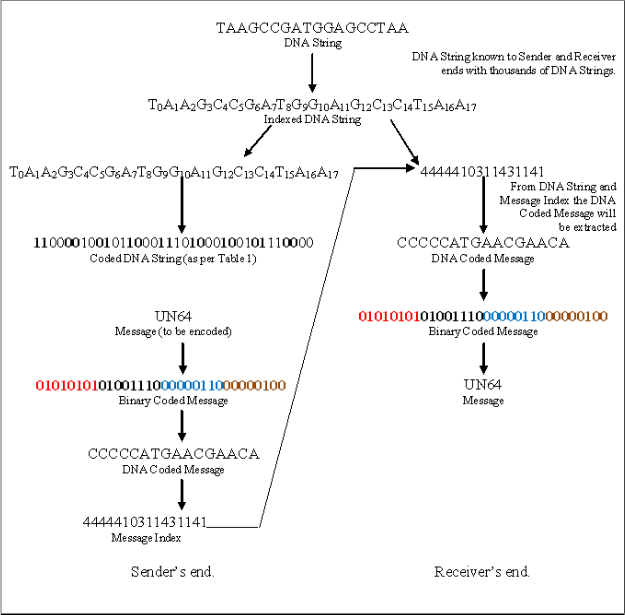
Fig. 2. Encoding and Decoding Schemes.
Decoding message from DNA String and Message Index.
Step 1: Message Index, 4444410311431141 mapped with DNA
String, TAAGCCGATGGAGCCTAA => T0A1A2G3C4C5G6A7T8G9G10A11G12C13C14TA15A16
Step 2: Extracted DNA String,
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 4, Issue 2, February-2013 4
ISSN 2229-5518
CCCCCATGAACGAACA
Step 3: DNA Coded Message from Extracted DNA String. CCCCCATGAACGAACA =>
01010101010011100000011000000100
Step 4: Binary Coded Message from DNA Coded Message.
01010101010011100000011000000100 =>
01010101010011100000011000000100
Step 5: Computed Text (Alphanumeric) value is
01010101010011100000011000000100 => UN64
4 RESULT AND COMPARISION
If length of DNA Sequence is 100 characters with 8 characters are encoded then 1008 or 10 quadrillion (10,000,000,000,000,000), possible combinations are possible. If the system h as a built-in delay of only 0.01 second following the selection of each Characters until the end of the String or Sequence, it would take (on average) millions of years to break. Our work is explained in detail in Fig. 2.
Mohammad Reza Abbasy, et al, in 2011, claimed the proba- bility of making successful guesses of an attacker was as fol- low [2]:
[5] Boris Shimanovsky, Jessica Feng, and Miodrag Potkonjak, “Hiding Data in DNA”, Workshop on Information Hiding, Noordwijkerhout, The Nether- lands, October 7-9, (2002), pp. 373-386.
[6] [http://dnasequencing.com/ [Last accessed on February 13, 2012]
[7] http://www.ebi.ac.uk/ [Last accessed on February 13, 2012]
1 1 1
------------ X ----- X -----
163 x 106 24 24
5 CONCLUSION
Hiding data in DNA Sequence is a very new concept. As per our study and discussions of our algorithms, it is very difficult to break and guess actually embedded data from the se- quence. There are millions of DNA Sequences are available according to European Bioinformatics Institute (EBI) [7].
This scheme may be questionable if biological mutation of Gene taken place in the middle. So, there is a scope for further study works on this line.
REFERENCES
[1] Debnath Bhattacharyya, P. Das, S. Mukherjee, D. Ganguly, S.K. Bandyo- padhyay, Tai-hoon Kim, “A Secured Technique for Image Data Hiding”, Communications in Computer and Information Science, Springer, June, (2009), Vol. 29, pp. 151-159.
[2] Mohammad Reza Abbasy, Azizah Abdul Manaf, and M.A. Shahidan,
“Data Hiding Method Based on DNA Basic Characteristics”, International
Conference on Digital Enterprise and Information Systems, July 20-22, (2011), London, UK, pp. 53–62.
[3] Mohammad Reza Najaf Torkaman, Pourya Nikfard, Nazanin Sadat Kaza- zi, Mohammad Reza Abbasy, and S. Farzaneh Tabatabaiee, “Improving Hybrid Cryptosystems with DNA Steganography”, International Confer- ence on Digital Enterprise and Information Systems, July 20-22, (2011), London, UK, pp. 42-52.
[4] Dominik Heider and Angelika Barnekow, “DNA watermarks: A proof of concept”, BMC Molecular Biology, April 21, (2008), Vol. 9, Issue 40, pp. 1-
10. (http://www.biomedcentral.com/1471-2199/9/40)
IJSER © 2013 http://www.ijser.org


