
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1272
ISSN 2229-5518
Authors: - Dr Yagnesh Thakar, Miss Neha N Singh
Abstract
Blood culture is a microbiological culture of blood. It is employed to detect infections that are spreading through the bloodstream (such as bacteremia, septicemia amongst others). The blood culture represents a critical tool for the health care professional as a means of detecting the dangerous presence of living organism in the blood stream. A positive blood culture can suggest a definitive diagnosis, enable the targeting of therapy against the specific organisms and provide prognostic value (Bryan C.S 1989). In blood culture, false positive arises due to contamination which occurs when organisms that are not actually present in a blood sample are grown in culture.
Faced with a positive blood culture result, clinicians must determine whether the organism represents a clinically significant infection associated with great risk of morbidity and mortality. Further complicating the issue in recent years is the increasing use of central venous catheters (CVC) and other indwelling vascular access devices (Anonymous 1986, Data summary from October 1986- April1996, issued in May
1996)( National Nosocomial Infections surveillance system 2004, Data summary from January 1992 through June 2004, issued October 2004)
Interpretation of culture result for patients with these devices in place is particularly challenging because while these individuals are at increased risk for bacteremia, such results may also indicate culture contamination or colonization of the line. It has been demonstrated in a number of studies of both adult and pediatric patients (Bates, D.W; Goldman and T.H. Lee 1991)(Segal, G.S; and J.M Chamberlain
2000)(Souvenir, D;D.E. Anderson, Jr; S Polpant, H Mroch, S.Askin, J.Anderson, J.Claridge, J. Eiland, C. Malone, M.W Garrison, P.Watson, and D.M Campbell1998) .
The college of American pathologists (CAP) Q Probes quality improvement study involved the prospective examination of 497,134 blood culture specimen from 640 U.S health care institutes (Schifman, R.B and A. Pindur 1993). While for other, it has more than 5% of their blood culture were contaminated. The financial impact of blood culture contamination has been described in a number of studies (Bates, D.W; L. Goldman and T.H Lee 1991) (Dunagan, W.C; R.S Woodward, G. Medoff, J.L Gray III, E. Casabar, M.D Smith, C.A Lawrenz and E. Spitznage L. 1989)(Waltzman, M.L; and M. Harper 2001)
Bates et al. found that contaminant results, compared with true negative result, were independently associated with increased subsequent charges and intravenous antibiotic charges (Bates D.W; L.Goldman and T.H Lee 1991). In a subsequent study focused on blood culture contamination caused by coagulase negative staphylococci, Souvenir et.al reported that almost half of the patients with a false positive result were treated with antibiotics, often with Vancomycin (Souvenir D; D.E Anderson, Jr; S Palpant, H Mroch, S. Askin, J. Anderson, J. Claridge, J. Eiland, C. Malone, M.W. Garrison, P.Watson and D.M Campbell 1998).
In a retrospective study of 9,959 blood cultures performed in children aged 1 month to 18 years, Thuleret. Al found that 26% of children with false positive culture who were initially evaluated as outpatients were subsequently admitted to the hospital on the basis of initial culture results (Thuler, L.C; M. Jenicek, J.P Turgeon, M.Rivard, P. Lebel and M.H Lebel 1997). The presence of living microorganisms in blood has substantial clinical importance. From the diagnostic standpoint a positive blood culture yielding a clinically important microorganism represents either failure of host defences to contain an infection at its primary focus. A positive blood culture however is not always clinically significant, since contamination may occur or the positive result may represent the presence of microorganisms in the blood (Everett E.D, Hirschmann IV
1997).
A Comprehensive analysis of positive blood culture in 1990s, authored by two of the current investigators (Weinstein M.P, Reller L.B, Murphy
J.R, Lichtenstein K.A, 1983). The primary goal is to determine the current microbiology, epidemiology and importance of positive blood culture, with special references to similarities and differences (Gold, Polsky 1986).
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1273
ISSN 2229-5518
Identification of organisms:
Bates et. al. found that the identification of organisms was the most important predictor for differentiate the contaminated blood culture results from indicating bacteremia or septicemia (Bates D.W; and T.H Lee 1991)
Weinstein et.al study positive blood culture in adult inpatients from 3 hospitals around the country suggested that certain organisms should almost always be thought to represent true septicemia or fungemia when isolated from a blood culture (Weinstein, M.P; M.L Towns, S.M Quartey, S.Mirrett, L.G Reimer, G. Parmigiani and L.B Reller 1997). These organisms included Staphylococcus aureus, S.pneumoniae, E.coli and other Enterobacteriaceae, Pseudomonas aeruginosa and candida albicans. Weinstein’s personal observation is that the following organisms always represent a true infection when isolated fron blood culture, Streptococcus pyogenes, S.agalactiae, Haemophilius influenza, members of all bacteriology fragilis group, all candida species and Cryptococcus neoformans (Weinstein, M.P 2003)
Along the same vein, certain organisms have been found to represent contamination in a significant proportion of cases. These organisms include coagulase negative staphylococci, Bacillus species, Micrococcus species, Streptococci. Enterococci and clostridium perfingens (Weinstein M.P; M.L Towns, S.M Quartey, S. Mirrett, L.G Reimer, G.Parmigini and L.B Reller 1997) .However, it is crucial to recognize that each of these organisms can also represent true bacteremia or septicemias. Of these organisms, the ones that are thought to represent true bacteremia or septicemia are corynebacterium species, Bacillus species and other B. Anthracis (Weinstein M.P 2003).
Coagulase negative staphylococci were usually believed to represent contamination when isolated from blood cultures. Hence it is the most common blood culture contaminants which represents 70% to 80% of all contaminated blood cultures (Calfee, D.P; and B.M Farr
2002)(Norberg, A.N.C.Christopher, M.L Ramunodo, J.R Bower and S.A Berman 2003)(Rubin L.G; P.J Sanchez, J.Siegel, G. Levine, L.Saimann 2002)
Weinstein et al found that even though only 12.4% of coagulase negative staphylococcal isolates were clinically significant, they ranked as the third most common cause of septicemia. Similarly, other organisms can be difficult to interpret when isolated from blood cultures. One study found that enterococci were pathogens 70% of the time, whereas viridians group streptococci were pathogens 38% of the time (Weinstein, M.P; M.L Towns, S.M Quarty, S.Mirrett, L.G Reimer, G.Parmigiani and L.B Reller 1997). Clostridium perfringens was contaminants 77% of the time, whereas other clostridium species were true pathogens 80% of the time. Bloodstream infections involve only a single organism, prompting clinicians to sometime conclude that a blood culture bottle that grows multiple organisms is contaminated. However, studies have found that 6% to 21% of all true bacteremia are polymicrobial, usually in patients in high risk group (Sharma, M.K, Riederer, L.B Johnson ans R. Khatib
2001).
Multiple coagulase negative species have been found to cause polyclonal coagulase negative staphylococci infections (Galdbart, J.O; A.Morvan, (N.Desplace and N.elSolh 1999) (Van Eldere, J; W.E Peetermans, S.VanLierde and J. Van Eldere, 1997).
Number of positive blood culture bottles within a blood culture sets:
Another method that has been used by health care workers to differentiate contaminated blood cultures from culture that represent septicemia in the number of blood culture bottles that exhibit growth within a given blood culture set. Theoretically, if one bottle exhibits growth within a given set. The likelihood of contamination is greater. However, there is at least one study that suggested that these criteria should not be used for this purpose (Mirrett S; L.B Reller, C.A Petti, C.W Woods, M.L Wilson, M.P Weinstein 2001).
In the study, Mirrett et al. found that the number of bottles positive for coagulase negative staphylococci within blood culture sets comprising two, three or four bottles was not correlated with the likelihood of infections using clinical parameters( Mirrett S; L.B Reller, C.A
Petti, C.W Woods, M.L Wilson, M.P Weinsten 2001)
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1274
ISSN 2229-5518
Usually, a set of blood cultures involve one aerobic bottle and one anaerobic bottle in an attempt to optimize the isolation of both aerobic and anaerobic organisms. It makes sense intuitively that if growth of a given organisms is more likely in aerobic conditions than anaerobic conditions, the number of positive culture bottles within a set that consist of one aerobic conditions and one anaerobic bottle should not be used to differentiate contaminated from clinically significant cultures. Recent studies have found that the presence of growth for a given organism varies between the aerobic and anaerobic bottle. One study of 644 positive blood culture sets found that 413(59.8%) were recovered from both bottle, 206(29.8%) were recovered only from aerobic bottle and 2(10.4%) were recovered only from anaerobic bottle (Saito, T; K Senda, S. Takaura, N. Fujihara, T. Kudo, Y.Linuma, M.Tanimoto and S.I schiyama 2003).
In that study, the aerobic bottle was significantly superior to the anaerobic bottle for both recovery and detection time for overall organisms and there was no significant difference in detection time for facultative anaerobic bacteria between the two bottles. Another study of coagulase negative staphylococcal isolates found that the majority (59.7%) of isolates grew in the aerobic bottle only, whereas 27.7% grew in both bottles and 12.5% grew only in the anaerobic bottle (Khatib, R; K.M Riederer, J.A Clark, S. Khatib, L.E Briski and F.M Wilson 1995). Another factor in determining contamination that has been explored by several investigators is the amount of time required for the organism to grow in the culture medium. It is thought that perhaps the blood from septicemic patients will have a much higher inoculum of bacteria than a contaminated culture. Theoretically, it follows that a larger inoculum will grow faster than a small inoculum, a theory that seems to have verified in prior studies of catheter related bloodstream infections (Catton; J.A; B.M Dubbins, P. Kite, J.M Wood, K.Eastwood, S. Sugden, J.A Sandae, D. Burke, M.J Mc Mahon and M.H Wilcox 2005)( Frankin J.A; A.H Gaur, J.L Shenep, X.J Hu and P.M Flynn 2004)( Innes, G.K Roland, E. Grafstein and J.M Christenson 2000)( Rogers, M.S; and B.A Oppenhein 1998).
In support of this theory, several studies have shown that cultures that become positive more than 3 to 5 days after incubation have been more likely to represent contamination (Evans, M.R; A.L Truant, J. Kostmann and L. Locke 1991)(Hans. X.Y; and A.L Truant
1999)(Herwaldt, L.A; B.B Hulbert and P.C Migneault 1992)(Huang. A.H; J.J Yan and J.J Wee 1998)(Kurlat I; B.J Stoll and J.E Mac Gowan Jr.
1989)(Mozes B; L.B Reller; C.A Petti; C.W Woods, B. Vaiziran, R. Sivadas and M.P Weinstein 2003). Because it can be difficult to obtain more than one set of cultures in the pediatric populations.
Haimi- Cohen et al used clinical parameters to differentiate true coagulase negative infections from contaminants and found that a time to positively of <15 hr had a positive predictive value of 84% for true infection in children (Haimi- Cohen, Y;S Shaffnoori, V. Tucci and L.G Rubin 2003)
Bates et al found that the time to growth was a useful variable in a multivariable algorithm for predicting true septicemia from a positive culture results, although it did not perform as well as either the identification of the organism or the presence of multiple positive cultures (Bates, D.W; and T.H Lee 1995). Souvenier et al study which differentiated coagulase negative staphylococcus contaminated cultures from septicemia or bacteremia in the time for detection of a positive culture.
Khatib et al’s molecular study of 47 episodes of multiple positive culture for coagulase negative staphylococci found that time to growth does not help differentiate cultures that grew identical strains from culture sets that differed by strain. The difference in time to detect the contaminants versus true positive coagulase negative staphylococci was statically significant. The time to growth and sensitivity for detecting growth can be expected to change, making the use of this technology in this regard questionable.
Population studies:
In the pediatric population, investigators have studied the risk of occult bacteremia, given the widespread adoption of vaccinations to prevent infection caused by Haemophilus Influenza and streptococcus pneumonia (Alpern, E.R; E.A Alessandrini, L.M Bell, K.N Shaw and K.L;
McGowan 2000)(Bandyopadhyay, S; J. Bergholte, C.D Blackwell, J.R Friedlander and H. Hennes 2002). In a study of well appearing, highly
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1275
ISSN 2229-5518
febrile children aged 2 to 36 months, Stoll and Rubin found that the incidence of occult bacteremia was 0.91% and concluded that universal blood culture testing in this population may be unnecessary.
Herz et al conducted a retrospective review of blood cultures obtained fron children aged 3 to 36 months during a five year period from 1998 to
2003. Bacteremia rates dropped from 1.62% to 0.71% from the first year to the last year of the study period, while the contaminant to pathogen ratio increased from 1:2:1 to 2:3:1, leading the authors to agree with Stoll and Rubin about the decreased role for blood culture testing in highly febrile children in this age group. Innes et al found that during a 6 month period at a hospital, blood culture result from patients seen in the emergency department were rarely helpful. With only 2.1% of 767 cultures yielding potentially helpful, with only 2.1% of retrospective study of
1,350 patients who had blood cultures taken in the emergency department, only 0.52% had results that potentially affected their management.
Contaminated blood cultures are a particular challenge for infants and children for several reasons. Evidence suggests that contamination occurs more frequently in this population, particularly in young infants. In addition, concerns about the risk of occult bacteremia it have led to use blood culture and empirical therapy, particularly in children less than 3 year of age (Baraff, L. J; J.W Bass. G.R Fleisher, J.O Klein, G.H McGacken, Jr; K.R Powell, D.L; Schriger, et al 1993). The current era of influenza and pneumococcal vaccination, the risk of occult bacteremia has significantly lowered. As a result, the use of blood culture testing on this patient’s population is associated with a lower positive predictive value. Moreover, analysis of current practice patterns reveals that in most cases, only single blood cultures are collected. In an effort to reduce unnecessary discomfort, pediatricians often use existing intravenous catheters for obtaining cultures instead of peripheral venipuncture.
Ramsook et al found that similar results in a 6 month study of 2,431 pediatric blood cultures with contamination rates of 3.4% for specimen collected via intravenous catheters versus 2.0% for those obtained by separate venipuncture. Ramsook single blood cultures are particularly common in pediatric patients combined with the increased utilization of catheter based culture contamination challenging, particularly when coagulase negative staphylococci are grown in culture (Ammann, R.A; A. Hirt, A.R Luthy and C. Aebi 2003) .
Neonatal septicemia is an important cause of morbidity and mortality among neonates in India. With an estimated incidence of approximately 4% in intramural livebirths (Indian Pediater 1997). Delayed treatment until clinical recognition of signs and symptoms of sepsis entails risk of preventable mortality, presumptive antibiotic therapy may result in overtreatment. A wide variety of bacteria both aerobic and anaerobic can cause neonatal septicemia. Blood culture was done for all neonates suspected to have septicemia.
All blood culture was collected from a peripheral vein with proper aspectic precautions before starting any antibiotic therapy. Approximately 3 ml of blood was inoculated into brain heart infusion broth and incubated at 37 c. subcultures were made on both blood and Mac conkey agar after 24 and 48 hrs. Growth if any, was identified by the standard bacteriological techniques ( Cruickshank k, Duguid J P, Marmion BP, Swain RHA, Livingstone 1975), including gram staining, colony characteristics, biochemical properties. Antibiotic sensitivity was performed by Stroke’s Disc Diffusion method (Strokes EJ, Arnold 1975).
Antimicrobial susceptibility Test:
The result of direct antimicrobial susceptibility testing of positive blood cultures have been shown to correlate well with those of standardized tests. In most of the foregoing studies, blood culture samples were subculture to broth to allow the adjustment of the inocula before susceptibility testing (Johnson, J.E; and J.A Washington III 1976) (Kiehn, T.E; C. Capitolo and D. Armstrong 1982) (Mirrett, S; and L. BarathReller 1979) (Wegner, D.L; C.R Mathis and T.R Neblett 1979).
Reller et al recommend that the inoculum density should be adjusted to match a 0.5 Mcfarland turbidity standard for direct susceptibility tests for blood cultures (Reller. L.B; P.R Murray and J.D Maclowry 1982). It is common practice in many laboratories to report presumptive
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1276
ISSN 2229-5518
susceptibilities after 4 to 6 hr incubation of direct disk diffusion tests from unadjusted blood cultures. This study was untaken to test the accuracy of 4 to 6 hr readings of disk diffusion tests that were inoculated directly from positive blood cultures.
Arbo et al found that blood cultures have a limited effect on antibiotic choices as they are observed that there was underutilization of the culture and sensitivity results. The most common used antibiotic was Quinolone particularly Ciprofloxacin (39%) followed by Chloramphenicol (22%) and Co- amoxiclav (16%). Monotherapy was also started in 50% cases of secondary bacteremia, the combinations of Clindamycin+ an aminoglycoside and Ciprofloxacin + Aminoglycoside and Ciprofloxacin + Aminoglycoside were the preferred antibiotic. Evaluation of the antibiotic regimen given showed only 7 out of 29 or 24.1% were used. This is comparable to the study of Edwards Id, el al 1973, Arbo MDJ
1994, Kunin CM, Tupasi TE, Craig WA 1973 and Castle M et al 1977.
In the study of Arbo, modification of antibiotic therapy consisted of addition of antibiotics. This is also true in our study, wherein
48% of the antibiotic therapy is superfluous. Both the studies of Kunin and Castle showed less than 50% of approximately Antibiotic use. In neonates, Vancomycin is still the drug of choice of S.aureus but resistance to this drug has also been reported (Jick S. 1997). A combination of Ciprofloxacin and Amikacin appears to be the best choice for infections due to Klebsiella. These findings are in tandem with the National Neonatal Perinatal Database.
Treatment with Ciprofloxacin is also indicated in multidrug resistant S.aureus in the pediatrics age group, but its use in neonates is still experimental due to lack of safety data (Karthikeyan G, Premkumar K 2001, Jick S 1997).
Automated blood culture system:
The advent of automated blood culture detection systems allows significantly earlier detection of most aerobic bloodstream pathogens than manual systems. (Rohner P et al 1995)
Introduction
Septicemia is the presence of bacteria in the blood (bacteremia) and is often associated with severe disease. Its alternative name is blood poisoning (bacteremia with sepsis).Septicemia is a serious, life-threatening infection that gets worse very quickly. It can arise from infections throughout the body, including infections in the lungs, abdomen, and urinary tract. It may come before or at the same time as infections of the bone (osteomyelitis), central nervous system
(meningitis), or other tissues. Sepsis is a
severe illness caused by overwhelming infection of the bloodstream by toxin- producing bacteria.Its alternative name Systemic inflammatory response syndrome (SIRS) .Sepsis is caused by bacterial infection that can originate anywhere in the body. Blood cultures should be obtained (PRIOR to initiation of antimicrobial therapy) for any patient, in whom there is suspicion of bacteremia, including hospitalized patients with fever and leukocytosis or leukopenia. Circumstances in which blood cultures are especially
important include sepsis, meningitis,
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1277
ISSN 2229-5518
osteomyelitis, arthritis, endocarditis, pneumonia, and fever of unknown origin.
The infection is often confirmed by a positive blood culture, though blood cultures may be negative in individuals who have been receiving antibiotics. In sepsis, blood pressure drops, resulting in shock. Major organs and systems, including the kidneys, liver, lungs, and central nervous system, stop functioning normally. Systemic inflammatory response syndrome or SIRS is evidence of the body's ongoing inflammatory response. When SIRS is suspected or known to be caused by an infection, this is sepsis. Severe sepsis occurs when sepsis leads to organ dysfunction, such as trouble breathing, coagulation or other blood abnormalities, decreased urine production, or altered mental status. If the organ dysfunction of severe sepsis is low blood pressure (hypotension), or insufficient blood flow (hypoperfusion) to one or more organs (causing, for example, lactic acidosis), this is septic shock.Even though bacteremia and sepsis are closely
related, they are two separate conditions. The simple presence of bacteria in the blood is known as bacteremia. It may be transient,
where small quantities of bacteria are in the
blood for a limited period of time, or it can be sustained, where the bacteria persist and multiply in the bloodstream. The sustained form of bacteremia is usually what leads to sepsis, which is the body's immune response to the presence of the bacteria. This potentially fatal condition, sometimes referred to as blood poisoning, involves an inflammatory response by the entire body and is characterized by increased body temperature, heart rate, and respiratory rate, and in its severe form can lead to organ failure, extreme low blood pressure, or septic shock.
Sepsis can lead to multiple organ dysfunction syndrome (MODS) (formerly known as multiple organ failure), and death. Organ dysfunction results from local changes in blood flow, from sepsis-induced hypotension (< 90 mmHg or a reduction of ≥
40 mmHg from baseline) and from diffuse intravascular coagulation, among other things. Sepsis can be defined as the body's response to an infection. An infection is caused by microorganisms or bacteria invading the body and can be limited to a particular body region or can be widespread in the bloodstream. Sepsis is acquired quickest with infections developed in surgery and physical contact with someone
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1278
ISSN 2229-5518
with sepsis. Just over one-third of sepsis patients have positive blood cultures, mainly due to inadequate sampling volumes (50% of adults have < 1.0 CFU/mL blood) and the prior use of antibiotics. However, 20-30% of sepsis patients are given inappropriate empirical antibiotics. The Clinical and Laboratory Standards Institute guidelines recommend paired culture sets to help discriminate between contaminant organisms and true pathogens; four 10-mL bottles (2 sets) should be used for the initial evaluation to detect about 90-95% of bacteremias and six 10-mL bottles (3 sets) should be used to detect about 95-99% of bacteremias. It has also been shown that the positivity rate increased by 15-35% with resin-based media in patients on antibiotics. For diagnosing catheter-related bloodstream infections, differential time-to-positivity is one method recommended to help determine whether the catheter is truly the source of infection. The proper training of personnel with regard to drawing an appropriate blood volume and the importance of clear labeling of culture bottles is also of critical importance. Furthermore, if the contamination rate is relatively high, hiring dedicated staffs that are well-trained in order to get a lower blood culture contamination
rate may be cost-effective. It is because high
false-positive blood culture rates due to contamination are associated with significantly increased hospital and laboratory charges.
Bacteremia is the presence of viable bacteria in the bloodstream. Likewise, the terms viremia and fungemia simply refer to viruses and fungi in the bloodstream. These terms say nothing about the consequences this has on the body. For example, bacteria can be introduced into the bloodstream during tooth brushing (Lockhart, P. B.; Brennan, M. T.; Sasser, H. C.; Fox, P. C.; Paster, B. J.; Bahrani-Mougeot, F. K. (2008) This form of bacteremia almost never causes problems in normal individuals. However, bacteremia associated with certain dental procedures can cause bacterial infection of the heart valves (known as endocarditis) in high-risk patients(Wilson, W.; Taubert, K. A.; Gewitz, M.; Lockhart, P. B.; Baddour, L. M.; Levison, M.; Bolger, A.; Cabell, C. H. et al (2007).Conversely, a systemic inflammatory response syndrome can occur in patients without the presence of infection, for example in those with burns, polytrauma, or the initial state in pancreatitis and chemical pneumonitis (Bone, R.; Balk, R.; Cerra, F.; Dellinger, R.; Fein, A.; Knaus, W.; Schein, R.; Sibbald, W. (1992).
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1279
ISSN 2229-5518
Blood cultures are used to detect the presence of bacteria or yeasts in the blood, to identify the microorganism(s) present, and to guide treatment. Two or more blood cultures are typically ordered and collected as consecutive samples. Often, a complete blood count (CBC) is ordered along with or prior to the blood culture to determine whether the person has an increased number of white blood cells, indicating a potential infection. Sometimes other testing is also performed, such as a chemistry panel to evaluate the health status of a person's organs, or a urine, sputum, or cerebrospinal fluid (CSF) culture to help identify the source of the original infection. This is especially true when a person has symptoms associated with a urinary tract infection, pneumonia, or meningitis.
It is important to eliminate all of the bacteria that are causing the problem. For some infections, several weeks of treatment are necessary. This is especially important if you have endocarditis, which requires weeks of antibiotic therapy to cure.Thebacteria or yeast must grow in the nutrient media before they can be detected and identified. Usually this happens within a couple of days, but in some cases and with some microorganisms it can take longer.
Furthermore, some microorganisms are
present in the blood in very small numbers. They must have a sufficient time to reproduce. Additional blood cultures may be drawn to determine if bacteria present in the culture are persistent in the blood stream (true pathogens). If they are not present in follow-up cultures, then bacteria from the skin may have contaminated the initial cultures. Additional blood cultures may also be drawn if you continue to have signs of sepsis but no microorganism is recovered from the first cultures collected and grow to quantities that can be detected.In general, patients with bacteremia are likely to have low quantities of bacteria in the blood, even in the setting of severe clinical symptoms. For this reason, multiple blood cultures, each containing large volumes of blood, are required to detect bacteremia. Prior to initiation of antimicrobial therapy, at least two sets of blood cultures taken from separate venipuncture sites should be obtained. The technique, number of cultures, and volume of blood are more important factors for detection of bacteremia than timing of culture collection; these are discussed further in the following sections.
Technique — Careful technique is important to avoid contamination of the blood culture media by normal skin flora during the
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1280
ISSN 2229-5518
process of collection. This is important because normal bacterial skin flora can cause systemic disease such as infective endocarditis, and in some circumstances blood culture contamination can make it difficult to distinguish between false positive results and true infection. Important measures to reduce contamination include effective disinfection of the venipuncture site and avoiding blood culture collection through existing intravenous lines.
The tourniquet should be applied and the vein palpated before disinfection of the venipuncture site. Thereafter the venipuncture site should be cleansed with 70 percent alcohol followed by 1 to 2 percent tincture of iodine or chlorohexidine. The disinfectant should be allowed to dry for one to two minutes before blood is aspirated. If further palpation of the vein is necessary after skin preparation, a sterile glove should be worn. Alcohol should be used to disinfect the septum of culture bottles after removal of their flip caps. Blood should be collected directly into culture bottles during the venipuncture procedure, rather than into transport tubes sent to the laboratory for subsequent transfer of blood into the culture
bottle.
Definitions for this study for the most part were identical to those of Weinstein et al. (WeinsteinMP, MurphyJR, RellerLB, LichtensteinKA 1983)
Only a few will be detailed here. New or modified definitions also will be detailed.
True septicemia vs. contamination. Each positive blood culture was assessed critically by one of the investigators, each an infectious disease physician. All isolates were categorized as true-positives, contaminants, or of unknown clinical significance. The categorical decision was made after the following factors were taken into account: the patient's clinical history, physical findings, body temperature at the time of the blood culture, leukocyte count and differential cell counts, number of positive blood cultures out of the total number performed, results of cultures of specimens from other sites, imaging results, histopathologic findings, and clinical course and response to therapy. If the clinical significance of the positive culture was not clear on the basis of the available information, the isolate was categorized as being of unknown significance.
Episode. An episode of bacteremia, fungemia, or mycobacteremia was defined by the first positive blood culture in a series
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1281
ISSN 2229-5518
or by any new positive blood culture result that occurred >48 hours after the previous positive result, unless it was clear to the investigator that the new positive culture was part of the same episode. For example, a positive blood culture for Staphylococcus aureus on the fifth day of antibiotic therapy for S. aureus endocarditis was considered part of the original bacteremic episode.
Community- vs. hospital-acquired septicemia. Each episode was classified as either community- or hospital-acquired septicemia, according to guidelines of the Centers for Disease Control and Prevention (GarnerJS, JarvisWR, EmoriTG, HoranTC, HughesJM 1988) Septicemia in nursing home patients with positive blood cultures was always classified as hospital-acquired, whereas that in patients transferred from other outside facilities was classified according to their total period of hospitalization at both institutions.
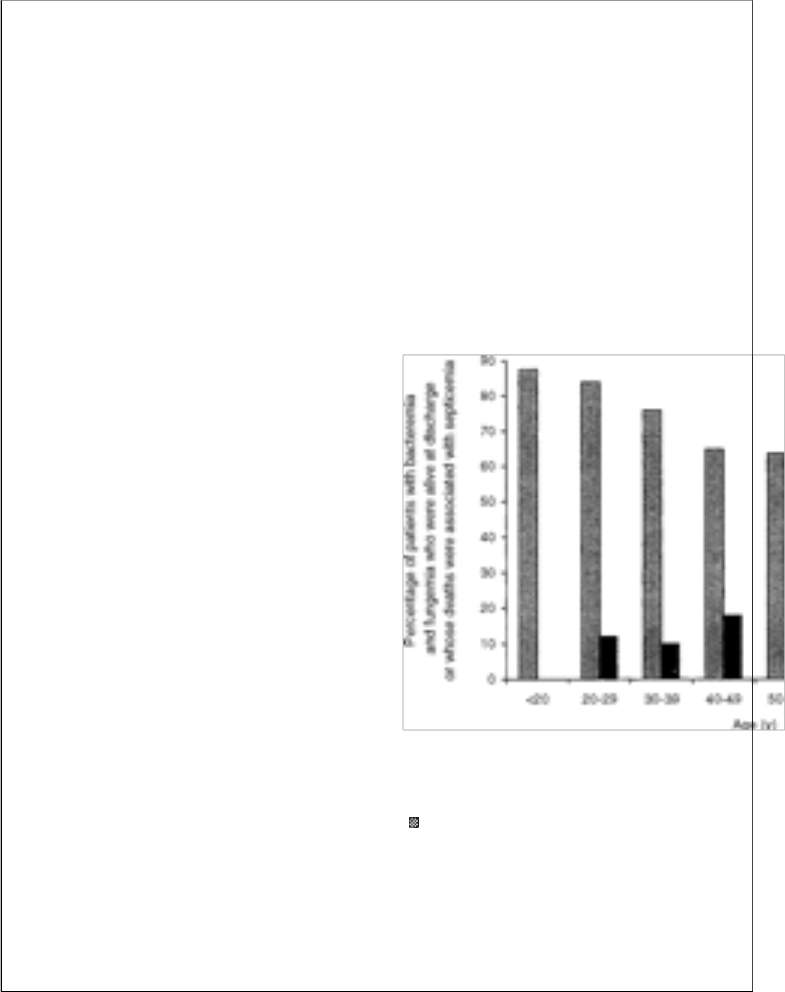
The effect of age on outcome of bacteremia and fungemia was assessed according to the decade of life of the infected patients. As shown in figure 1, septicemia-associated mortality gradually increased with age and was especially notable after the age of 70 years. The
infection-associated mortality for
septicemicpatients over 70 years of age was
24.2%, compared with 15.2% for patients 70 years of age or less. In comparison with other patients, the relative risks of death from bacteremia and fungemia for patients between 71 and 80 years of age and >80 years of age were 2.58 and 3.4, respectively. There were no significant differences in infection-associated mortality between septicemic men and women.
Figure 1. Outcome of bacteremia or fungemia, according to age group of patients ( = alive at discharge; ■ = death, associated with septicemia).
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1282
ISSN 2229-5518
Aims and Objectives
1) To study the profile of bacterial isolates from blood culture
2) To study antimicrobial resistance pattern of the isolates
3) To assess the advantages of
Automated blood culture
4) To study the frequency of blood culture positivity in clinically suspected cases of septicemias.
Review of Literature
Blood culture is a microbiological culture of blood. It is employed to detect infections that are spreading through the bloodstream (such as bacteremia, septicemia amongst others). The blood culture represents a critical tool for the health care professional as a means of detecting the dangerous presence of living organism in the blood stream. A positive blood culture can suggest a definitive diagnosis, enable the targeting of therapy against the specific organisms and provide prognostic value (Bryan C.S
1989). In blood culture, false positive arises due to contamination which occurs when organisms that are not actually present in a
blood sample are grown in culture.
Faced with a positive blood culture result, clinicians must determine whether the organism represents a clinically significant infection associated with great risk of morbidity and mortality. Further complicating the issue in recent years is the increasing use of central venous catheters (CVC) and other indwelling vascular access devices (Anonymous 1986, Data summary from October 1986- April1996, issued in May 1996)( National Nosocomial
Infections surveillance system 2004, Data
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1283
ISSN 2229-5518
summary from January 1992 through June
2004, issued October 2004)
Interpretation of culture result for patients with these devices in place is particularly challenging because while these individuals are at increased risk for bacteremia, such results may also indicate culture contamination or colonization of the line. It has been demonstrated in a number of studies of both adult and pediatric patients (Bates, D.W; Goldman and T.H. Lee 1991)(Segal, G.S; and J.M Chamberlain
2000)(Souvenir, D;D.E. Anderson, Jr; S Polpant, H Mroch, S.Askin, J.Anderson, J.Claridge, J. Eiland, C. Malone, M.W Garrison, P.Watson, and D.M Campbell1998) .
The college of American pathologists (CAP) Q Probes quality improvement study involved the prospective examination of
497,134 blood culture specimen from 640
U.S health care institutes (Schifman, R.B and A. Pindur 1993). While for other, it has more than 5% of their blood culture were contaminated. The financial impact of blood culture contamination has been described in a number of studies (Bates, D.W; L. Goldman and T.H Lee 1991) (Dunagan, W.C; R.S Woodward, G. Medoff, J.L Gray
III, E. Casabar, M.D Smith, C.A Lawrenz
and E. Spitznage L. 1989)(Waltzman, M.L;
and M. Harper 2001)
Bates et al. found that contaminant results, compared with true negative result, were independently associated with increased subsequent charges and intravenous antibiotic charges (Bates D.W; L.Goldman and T.H Lee 1991). In a subsequent study focused on blood culture contamination caused by coagulase negative staphylococci, Souvenir et.al reported that almost half of the patients with a false positive result were treated with antibiotics, often with Vancomycin (Souvenir D; D.E Anderson, Jr; S Palpant, H Mroch, S. Askin, J. Anderson, J. Claridge, J. Eiland, C. Malone, M.W. Garrison, P.Watson and D.M Campbell 1998).
In a retrospective study of 9,959 blood cultures performed in children aged 1 month to 18 years, Thuleret. Al found that 26% of children with false positive culture who were initially evaluated as outpatients were subsequently admitted to the hospital on the basis of initial culture results (Thuler, L.C; M. Jenicek, J.P Turgeon, M.Rivard, P. Lebel and M.H Lebel 1997). The presence of living microorganisms in blood has substantial clinical importance. From the
diagnostic standpoint a positive blood
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1284
ISSN 2229-5518
culture yielding a clinically important microorganism represents either failure of host defences to contain an infection at its primary focus. A positive blood culture however is not always clinically significant, since contamination may occur or the positive result may represent the presence of microorganisms in the blood (Everett E.D, Hirschmann IV 1997).
A Comprehensive analysis of positive blood culture in 1990s, authored by two of the current investigators (Weinstein M.P, Reller L.B, Murphy J.R, Lichtenstein K.A, 1983). The primary goal is to determine the current microbiology, epidemiology and importance of positive blood culture, with special references to similarities and differences (Gold, Polsky 1986).
Identification of organisms:
Bates et. al. found that the identification of organisms was the most important predictor for differentiate the contaminated blood culture results from indicating bacteremia or septicemia (Bates D.W; and T.H Lee 1991)
Weinstein et.al study positive blood culture in adult inpatients from 3 hospitals around the country suggested that certain organisms should almost always be thought
to represent true septicemia or fungemia
when isolated from a blood culture (Weinstein, M.P; M.L Towns, S.M Quartey, S.Mirrett, L.G Reimer, G. Parmigiani and L.B Reller 1997). These organisms included Staphylococcus aureus, S.pneumoniae, E.coli and other Enterobacteriaceae, Pseudomonas aeruginosa and candida albicans. Weinstein’s personal observation is that the following organisms always represent a true infection when isolated fron blood culture, Streptococcus pyogenes, S.agalactiae, Haemophilius influenza, members of all bacteriology fragilis group, all candida species and Cryptococcus neoformans (Weinstein, M.P 2003)
Along the same vein, certain organisms have been found to represent contamination in a significant proportion of cases. These organisms include coagulase negative staphylococci, Bacillus species, Micrococcus species, Streptococci. Enterococci and clostridium perfingens (Weinstein M.P; M.L Towns, S.M Quartey, S. Mirrett, L.G Reimer, G.Parmigini and L.B Reller 1997) .However, it is crucial to recognize that each of these organisms can also represent true bacteremia or septicemias. Of these organisms, the ones that are thought to represent true bacteremia or septicemia are corynebacterium species,
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1285
ISSN 2229-5518
Bacillus species and other B. Anthracis
(Weinstein M.P 2003).
Coagulase negative staphylococci were usually believed to represent contamination when isolated from blood cultures. Hence it is the most common blood culture contaminants which represents 70% to 80% of all contaminated blood cultures (Calfee, D.P; and B.M Farr 2002)(Norberg, A.N.C.Christopher, M.L Ramunodo, J.R Bower and S.A Berman 2003)(Rubin L.G; P.J Sanchez, J.Siegel, G. Levine, L.Saimann
2002)
Weinstein et al found that even though only 12.4% of coagulase negative staphylococcal isolates were clinically significant, they ranked as the third most common cause of septicemia. Similarly, other organisms can be difficult to interpret when isolated from blood cultures. One study found that enterococci were pathogens
70% of the time, whereas viridians group streptococci were pathogens 38% of the time (Weinstein, M.P; M.L Towns, S.M Quarty, S.Mirrett, L.G Reimer, G.Parmigiani and L.B Reller 1997). Clostridium perfringens was contaminants 77% of the time, whereas other clostridium species were true pathogens 80% of the time. Bloodstream infections involve only a single organism,
prompting clinicians to sometime conclude that a blood culture bottle that grows multiple organisms is contaminated. However, studies have found that 6% to
21% of all true bacteremia are polymicrobial, usually in patients in high risk group (Sharma, M.K, Riederer, L.B Johnson ans R. Khatib 2001).
Multiple coagulase negative species have been found to cause polyclonal coagulase negative staphylococci infections (Galdbart, J.O; A.Morvan, (N.Desplace and N.elSolh 1999) (Van Eldere, J; W.E Peetermans, S.VanLierde and J. Van Eldere,
1997).
Number of positive blood culture bottles within a blood culture sets:
Another method that has been used by health care workers to differentiate contaminated blood cultures from culture that represent septicemia in the number of blood culture bottles that exhibit growth within a given blood culture set. Theoretically, if one bottle exhibits growth within a given set. The likelihood of contamination is greater. However, there is at least one study that suggested that these criteria should not be used for this purpose (Mirrett S; L.B Reller, C.A Petti, C.W
Woods, M.L Wilson, M.P Weinstein 2001).
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1286
ISSN 2229-5518
In the study, Mirrett et al. found that the number of bottles positive for coagulase negative staphylococci within blood culture sets comprising two, three or four bottles was not correlated with the likelihood of infections using clinical parameters( Mirrett S; L.B Reller, C.A Petti, C.W Woods, M.L Wilson, M.P Weinsten 2001)
Usually, a set of blood cultures involve one aerobic bottle and one anaerobic bottle in an attempt to optimize the isolation of both aerobic and anaerobic organisms. It makes sense intuitively that if growth of a given organisms is more likely in aerobic conditions than anaerobic conditions, the number of positive culture bottles within a set that consist of one aerobic conditions and one anaerobic bottle should not be used to differentiate contaminated from clinically significant cultures. Recent studies have found that the presence of growth for a given organism varies between the aerobic and anaerobic bottle. One study of 644 positive blood culture sets found that
413(59.8%) were recovered from both bottle, 206(29.8%) were recovered only from aerobic bottle and 2(10.4%) were recovered only from anaerobic bottle (Saito, T; K Senda, S. Takaura, N. Fujihara, T. Kudo, Y.Linuma, M.Tanimoto and S.I
schiyama 2003).
In that study, the aerobic bottle was significantly superior to the anaerobic bottle for both recovery and detection time for overall organisms and there was no significant difference in detection time for facultative anaerobic bacteria between the two bottles. Another study of coagulase negative staphylococcal isolates found that the majority (59.7%) of isolates grew in the aerobic bottle only, whereas 27.7% grew in both bottles and 12.5% grew only in the anaerobic bottle (Khatib, R; K.M Riederer, J.A Clark, S. Khatib, L.E Briski and F.M Wilson 1995). Another factor in determining contamination that has been explored by several investigators is the amount of time required for the organism to grow in the culture medium. It is thought that perhaps the blood from septicemic patients will have a much higher inoculum of bacteria than a contaminated culture. Theoretically, it follows that a larger inoculum will grow faster than a small inoculum, a theory that seems to have verified in prior studies of catheter related bloodstream infections (Catton; J.A; B.M Dubbins, P. Kite, J.M Wood, K.Eastwood, S. Sugden, J.A Sandae, D. Burke, M.J Mc Mahon and M.H Wilcox
2005)( Frankin J.A; A.H Gaur, J.L Shenep, X.J Hu and P.M Flynn 2004)( Innes, G.K Roland, E. Grafstein and J.M Christenson
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1287
ISSN 2229-5518
2000)( Rogers, M.S; and B.A Oppenhein
1998).
In support of this theory, several studies have shown that cultures that become positive more than 3 to 5 days after incubation have been more likely to represent contamination (Evans, M.R; A.L Truant, J. Kostmann and L. Locke
1991)(Hans. X.Y; and A.L Truant
1999)(Herwaldt, L.A; B.B Hulbert and P.C Migneault 1992)(Huang. A.H; J.J Yan and J.J Wee 1998)(Kurlat I; B.J Stoll and J.E Mac Gowan Jr. 1989)(Mozes B; L.B Reller; C.A Petti; C.W Woods, B. Vaiziran, R. Sivadas and M.P Weinstein 2003). Because it can be difficult to obtain more than one set of cultures in the pediatric populations.
Haimi- Cohen et al used clinical parameters to differentiate true coagulase negative infections from contaminants and found that a time to positively of <15 hr had a positive predictive value of 84% for true infection in children (Haimi- Cohen, Y;S Shaffnoori, V. Tucci and L.G Rubin 2003)
Bates et al found that the time to growth was a useful variable in a multivariable algorithm for predicting true septicemia from a positive culture results, although it did not perform as well as either the
identification of the organism or the
presence of multiple positive cultures (Bates, D.W; and T.H Lee 1995). Souvenier et al study which differentiated coagulase negative staphylococcus contaminated cultures from septicemia or bacteremia in the time for detection of a positive culture.
Khatib et al’s molecular study of 47 episodes of multiple positive culture for coagulase negative staphylococci found that time to growth does not help differentiate cultures that grew identical strains from culture sets that differed by strain. The difference in time to detect the contaminants versus true positive coagulase negative staphylococci was statically significant. The time to growth and sensitivity for detecting growth can be expected to change, making the use of this technology in this regard questionable.
Population studies:
In the pediatric population, investigators have studied the risk of occult bacteremia, given the widespread adoption of vaccinations to prevent infection caused by Haemophilus Influenza and streptococcus pneumonia (Alpern, E.R; E.A Alessandrini, L.M Bell, K.N Shaw and K.L; McGowan
2000)(Bandyopadhyay, S; J. Bergholte, C.D Blackwell, J.R Friedlander and H. Hennes
2002). In a study of well appearing, highly
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1288
ISSN 2229-5518
febrile children aged 2 to 36 months, Stoll and Rubin found that the incidence of occult bacteremia was 0.91% and concluded that universal blood culture testing in this population may be unnecessary.
Herz et al conducted a retrospective review of blood cultures obtained fron children aged 3 to 36 months during a five year period from 1998 to 2003. Bacteremia rates dropped from 1.62% to 0.71% from the first year to the last year of the study period, while the contaminant to pathogen ratio increased from 1:2:1 to 2:3:1, leading the authors to agree with Stoll and Rubin about the decreased role for blood culture testing in highly febrile children in this age group. Innes et al found that during a 6 month period at a hospital, blood culture result from patients seen in the emergency department were rarely helpful. With only
2.1% of 767 cultures yielding potentially helpful, with only 2.1% of retrospective study of 1,350 patients who had blood cultures taken in the emergency department, only 0.52% had results that potentially affected their management.
Contaminated blood cultures are a particular challenge for infants and children for several reasons. Evidence suggests that contamination occurs more frequently in this
population, particularly in young infants. In addition, concerns about the risk of occult bacteremia it have led to use blood culture and empirical therapy, particularly in children less than 3 year of age (Baraff, L. J; J.W Bass. G.R Fleisher, J.O Klein, G.H McGacken, Jr; K.R Powell, D.L; Schriger, et al 1993). The current era of influenza and pneumococcal vaccination, the risk of occult bacteremia has significantly lowered. As a result, the use of blood culture testing on this patient’s population is associated with a lower positive predictive value. Moreover, analysis of current practice patterns reveals that in most cases, only single blood cultures are collected. In an effort to reduce unnecessary discomfort, pediatricians often use existing intravenous catheters for obtaining cultures instead of peripheral venipuncture.
Ramsook et al found that similar results in a
6 month study of 2,431 pediatric blood cultures with contamination rates of 3.4% for specimen collected via intravenous catheters versus 2.0% for those obtained by separate venipuncture. Ramsook single blood cultures are particularly common in pediatric patients combined with the increased utilization of catheter based culture contamination challenging,
particularly when coagulase negative
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1289
ISSN 2229-5518
staphylococci are grown in culture (Ammann, R.A; A. Hirt, A.R Luthy and C. Aebi 2003) .
Neonatal septicemia is an important cause of morbidity and mortality among neonates in India. With an estimated incidence of approximately 4% in intramural livebirths (Indian Pediater 1997). Delayed treatment until clinical recognition of signs and symptoms of sepsis entails risk of preventable mortality, presumptive antibiotic therapy may result in overtreatment. A wide variety of bacteria both aerobic and anaerobic can cause neonatal septicemia. Blood culture was done for all neonates suspected to have septicemia.
All blood culture was collected from a peripheral vein with proper aspectic precautions before starting any antibiotic therapy. Approximately 3 ml of blood was inoculated into brain heart infusion broth and incubated at 37 c. subcultures were made on both blood and Mac conkey agar after 24 and 48 hrs. Growth if any, was identified by the standard bacteriological techniques ( Cruickshank k, Duguid J P, Marmion BP, Swain RHA, Livingstone
1975), including gram staining, colony
characteristics, biochemical properties.
Antibiotic sensitivity was performed by Stroke’s Disc Diffusion method (Strokes EJ, Arnold 1975).
Antimicrobial susceptibility Test:
The result of direct antimicrobial susceptibility testing of positive blood cultures have been shown to correlate well with those of standardized tests. In most of the foregoing studies, blood culture samples were subculture to broth to allow the adjustment of the inocula before susceptibility testing (Johnson, J.E; and J.A Washington III 1976) (Kiehn, T.E; C. Capitolo and D. Armstrong 1982) (Mirrett, S; and L. BarathReller 1979) (Wegner, D.L; C.R Mathis and T.R Neblett 1979).
Reller et al recommend that the inoculum density should be adjusted to match a 0.5
Mcfarland turbidity standard for direct susceptibility tests for blood cultures (Reller. L.B; P.R Murray and J.D Maclowry 1982). It is common practice in many laboratories to report presumptive susceptibilities after 4 to 6 hr incubation of direct disk diffusion tests from unadjusted blood cultures. This study was untaken to test the accuracy of 4 to 6 hr readings of disk diffusion tests that were inoculated directly from positive blood cultures.
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1290
ISSN 2229-5518
Arbo et al found that blood cultures have a limited effect on antibiotic choices as they are observed that there was underutilization of the culture and sensitivity results. The most common used antibiotic was Quinolone particularly Ciprofloxacin (39%) followed by Chloramphenicol (22%) and Co- amoxiclav (16%). Monotherapy was also started in 50% cases of secondary bacteremia, the combinations of Clindamycin+ an aminoglycoside and Ciprofloxacin + Aminoglycoside and Ciprofloxacin + Aminoglycoside were the preferred antibiotic. Evaluation of the antibiotic regimen given showed only 7 out of 29 or 24.1% were used. This is comparable to the study of Edwards Id, el al
1973, Arbo MDJ 1994, Kunin CM, Tupasi
TE, Craig WA 1973 and Castle M et al
1977.
In the study of Arbo, modification of antibiotic therapy consisted of addition of antibiotics. This is also true in our study, wherein 48% of the antibiotic therapy is superfluous. Both the studies of Kunin and Castle showed less than 50% of approximately Antibiotic use. In neonates, Vancomycin is still the drug of choice of S.aureus but resistance to this drug has also been reported (Jick S. 1997). A combination
of Ciprofloxacin and Amikacin appears to
be the best choice for infections due to Klebsiella. These findings are in tandem with the National Neonatal Perinatal Database.
Treatment with Ciprofloxacin is also indicated in multidrug resistant S.aureus in the pediatrics age group, but its use in neonates is still experimental due to lack of safety data (Karthikeyan G, Premkumar K 2001, Jick S 1997).
Automated blood culture system:
The advent of automated blood culture detection systems allows significantly earlier detection of most aerobic bloodstream pathogens than manual systems. (Rohner P et al 1995)
MATERIALS AND METHOD
Place of study: - Microbiology Lab of Care hospital and Vishakha clinical microbiology laboratory.
Period of study: - From 2nd of January 2012 to 15th April 2012
Subject of study:-.Automated Blood culture for the detection of Septicemia.
• Beaker
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1291
ISSN 2229-5518
• Conical flask
• Test tube
• Petri plate
• Inoculating loop
• Forcep
• Measuring cylinder
• Blood culture bottle
• Incubator
• Bac T Alert 3D system
• Cabinet (Hood)
• Burner
• Slide
• Autoclave
• Digital balance
• Refrigerator
• Alcohol
• Methylene blue
• Crystal violet
• Saffranine
• Gram iodide
• Distilled water
• Muller Hinton agar
• Mac Conkey agar
• Blood agar
Methods:
1) Specimen collection for blood culture:
The skin of veinpuncture site was disinfected by applying 70% alcohol in water with 1% iodine for atleast 1 min and allow it to dry. 5-10 ml of blood was collected.
The collected blood was then inoculated directly in Bac T/ Alert blood bottles.
2) Blood culture:
Blood culture were carried out as per standard protocol (Krishna el at 2001, forbes B A et al 2002 &vandepitte T et al 2003)
Glucose broth was inoculated for 24 hr at 34- 35oc . A sterile culture usually showed a layer of sedimented red blood covered by a pale yellow transparent broth.
Growth was observed by-
A floccular deposit on top of the blood layer
Turbidity
Coagulation of the broth
Production of gas
Culture was done on blood agar and mac conkey agar after
24 hrs, 48 hrs, 72 hrs, and 5th day. If after 5th day, no growth was obtained, the sample was considered as negative. If it is
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1292
ISSN 2229-5518

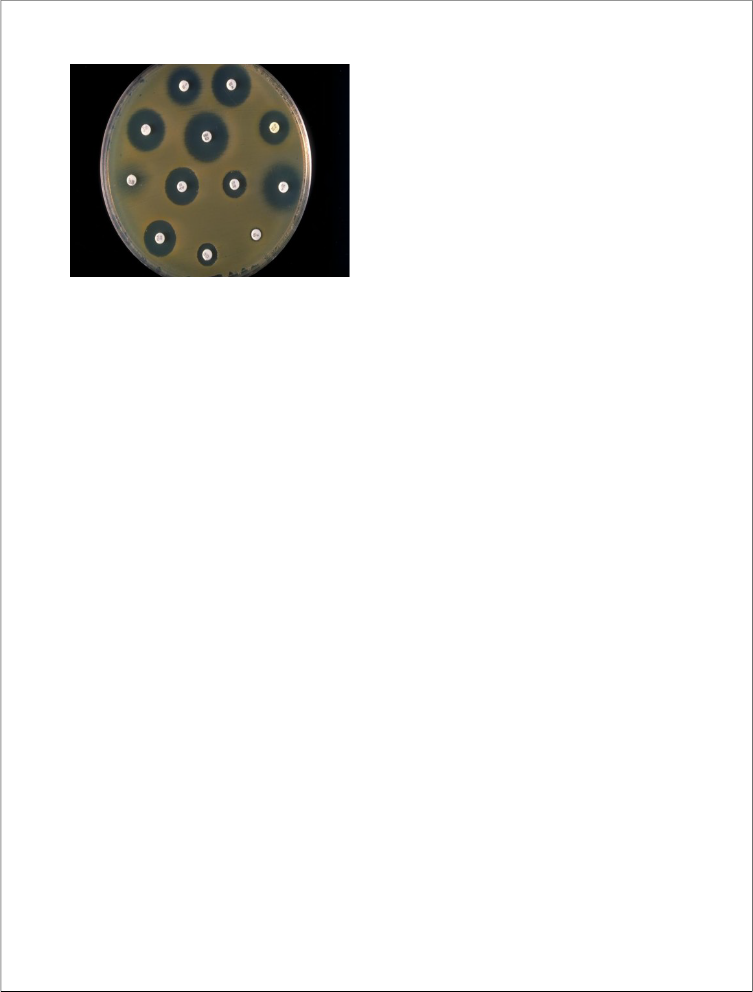
positive, colony morphology and standard biochemical tests were performed. (Refer Appendix 1,2)
3) Bac T/ Alert 3D System:
Automated blood culture system designed to do blood culture in both aerobic and anaerobic cultures. The capacity is 120 blood culture bottles.
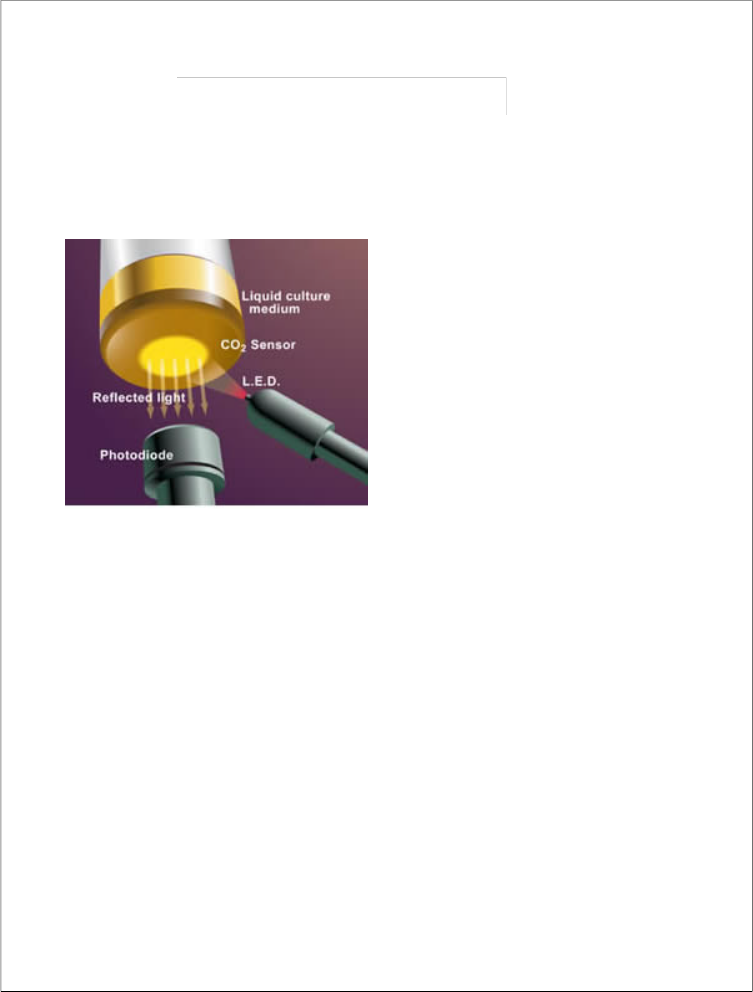
It is quick for the identification of bacteria grown in the bottle. It helps in quality control incorported and also in data management. 24 hrs continuous scanningof bottles were done for growth. The Power of Colorimetric Technology: Results are the most important aspect of any microbial detection system. Built on bioMérieux's patented colorimetric technology, the BacT/ALERT 3D demonstrates excellent recovery of a wide range of organisms with >95% recovery within
24 hours and >98% within 72 hours. BacT/ALERT Culture Media offer the ability to detect aerobic microorganisms and high-acid-producing organisms, such as lactobacillus, yeasts and molds. BacT/ALERT 3D's patented colorimetric sensor-and-detection technology detects
microorganism growth by tracking CO2
production. Notification of positives is immediate and results are dependable.
Working
1. Microorganisms multiply in the media, generating CO2 . As CO2 increases, the sensor in the bottle turns yellow.
2. Measuring reflected light, the BacT/ALERT 3D monitors and detects color changes in the sensor.
. Algorithms analyze the data to determine positivity, and the laboratory is notified immediately with visual and audible alarms.
4. Changes in the sensor are permanent and visible to the unaided eye, unlike any other method.
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1293
ISSN 2229-5518
such as shape, size, colony and nature of
5) Antimicrobial susceptibility test:
FAN)
Characterization of isolates:
Antimicrobial susceptibility testing was performed by Kirby – Bauer disc diffusion susceptibility method performed in accordance to NCCLS guidelines for gram positive and gram negative isolates.
Muller Hinton agar plate is used for susceptibility. (Refer appendix 3)
At least four morphologically similar colonies from an agar medium were touched with a wire loop and transferred to a tube containing 5 ml of nutrient broth. It is then incubated for 3-4 hrs. at
37oc to produce a suspension i.e.
turbidity. The density of suspension was matched with 0.5 MC farland standard.
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1294
IPSSoNp2u2l2a9-t5i5o1n8
Growth No
growth
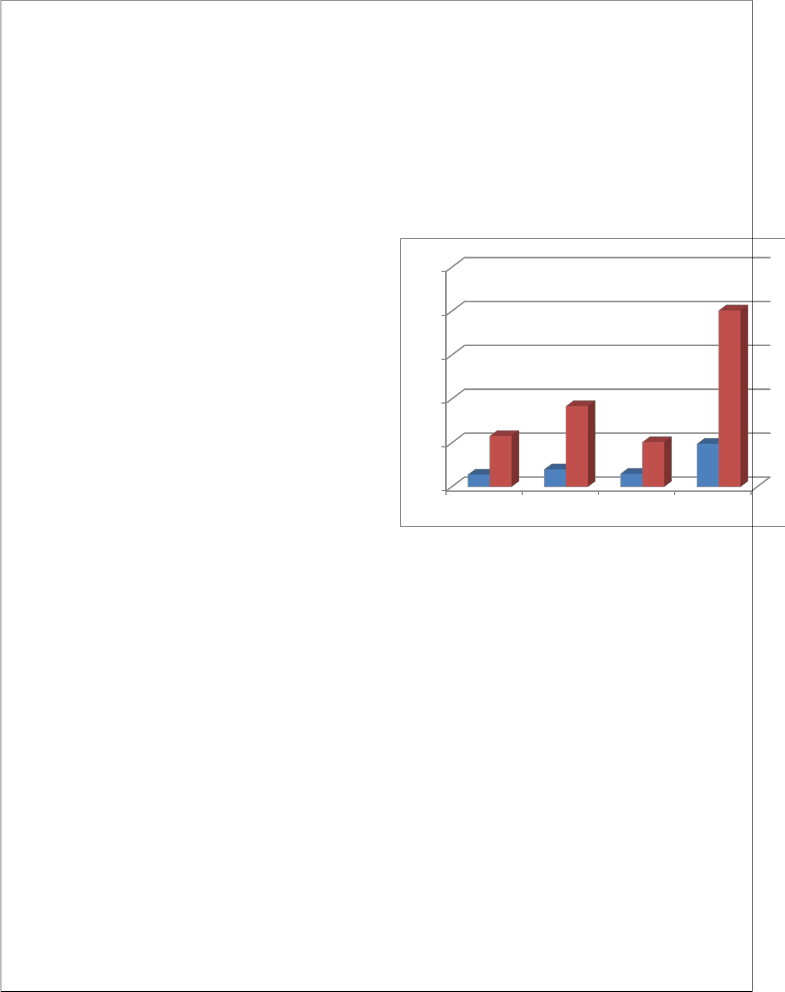
Adult 14 58 72
Children 20 92 112
Newborn 15 51 66
Total 49 201 250
Observation and results:
Table 1:
After that a sterile cotton swab was dipped into the suspension and extra fluid was removed by rotating the swab against the side of the tube. It is then streaked over the entire surface of the plate in four directions.
The antibiotic discs were applied to the agar surface with a sterile forceps and gently pressed down to ensure
contact. Commercially available discs
250
200
150
100
50
0
Adult Children New born Total
(Hi Media, Mumbai) with known potency were used. (Refer Appendix 4).
20 out of 112, and 92 were found to be negative in case of children. While new born shows positive culture 15 out of 66. And 51 were negative cultures in new born. From this table we can say that, the positivity arises more in children than in adult and new
born patients.
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1295
ISSN 2229-5518
Table 2:
Pseudomonas 11 22.4
Staphylococcus 20 40.8
Klebsiella 6 12.2
Acinetobacter 1 2.04
E.coli 8 16.3
Proteus 1 2.04
Candida 1 2.04
Salmonella typhi 1 2.04
22.4%, klesiella 12.2% and Acinetobacter, Proteus mirabilis, candida, and salmonella typhi shows 2.04%, while E.coli shows
16.3% positivity. Table 3:
Staphylococcus 3 10 7
Pseudomonas 5 5 1
E.coli 3 2 3
Klebsiella 2 1 3
Acinetobacter 0 1 0
Proteus
Mirabilis
Salmonella
Typhi
0 0 1
0 1 0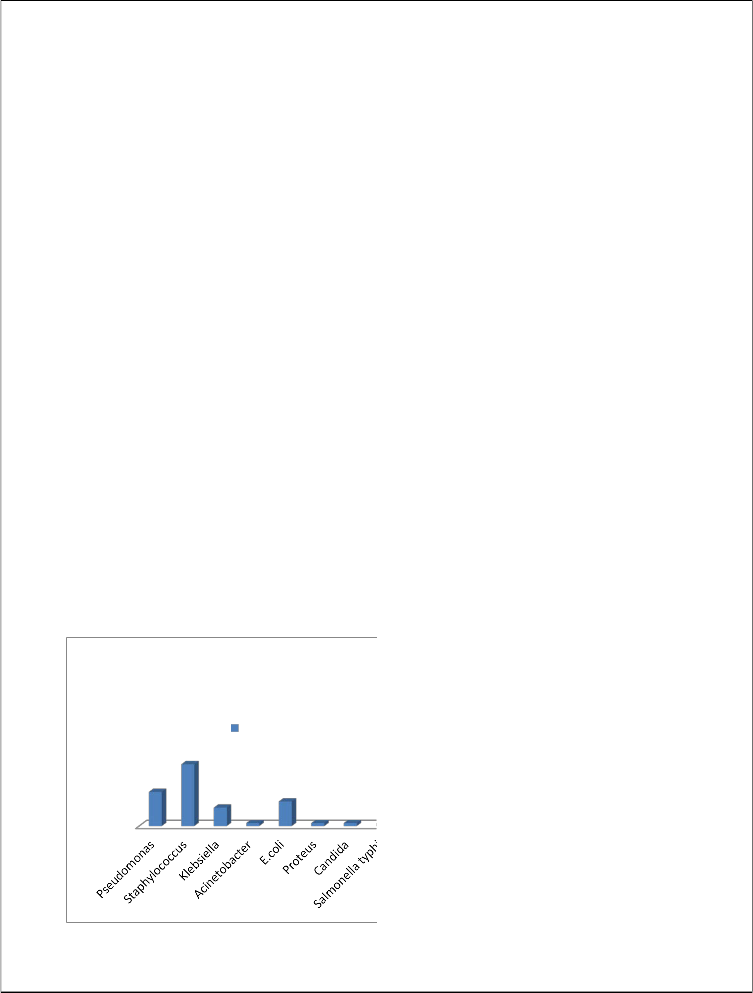
Number
20
11
6 8
1 1 1
Candida 1 0 0
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1296
ISSN 2229-5518
found to be more. In new born
Staphylococcus were found to be maximum. Table 4:
Klebsiella 6
E.coli 8
Proteus mirabilis 1
Acinetobacter 1
Salmonella typhi 1
Central line 8 1
Peripheral
line
234 48
blood culture in central line while in
peripheral line organisms like Pseudomonas,
Both 8 0 Klebsiella, E.coli, Proteus mirabilis, Acinetobacter, Salmonella typhi in
appropriate number.
The blood samples were taken from central line or peripheral line or both. Table 4 shows that central line blood culture shows less positivity than peripheral line. While central line+ peripheral line shows no positivity. Peripheral line shows 48 samples positive out of 234.
Table 5:
Table 6:
Antibiotic sensitivity for pseudomonas
I 10 90.9
MR 8 72.7
PT 5 45.5
AK 9 81.8
Sample Positive sample Organisms Total
G 10 90.9
Central line
1 Candida 1
LE 11 100
Peripheral
line
48 Pseudomonas
OF11
8 72.7
CF 7 63.6
Staphylococcus 20
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1297
ISSN 2229-5518
CPT 0 0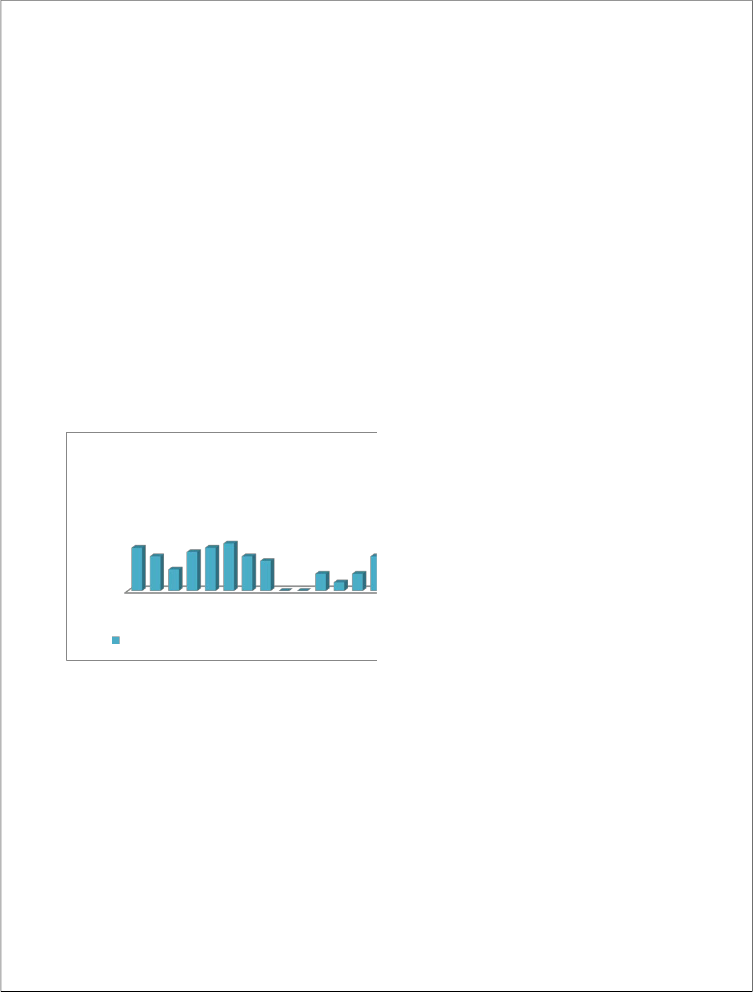
CA 0 0
CI 4 36.4
Antibiotic sensitivity for Staphylococcus
sensitivity for levofloxacin, while it is less
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1298
ISSN 2229-5518
CF 5 33.3
Antibiotic sensitivity for Staphylococcus N
18 17 17 18 18 19
CPT 3 20
CA 3 20
CI 5 33.3
10
2 6 6 4
12
3 5 1
CU 2 13.3
CFX 6 40
VA LZ G OF CD CA CU AC
Table 8:
Antibiotic sensitivity for GNB(E.coli, Klebsiella, proteus, samonella)
AC 8 53.3
A 5 33.3
CS 10 66.7
Antibiotic | No(n=15) | % |
I | 16 | 93.8 |
MR | 14 | 93.3 |
PT | 13 | 86.7 |
AK | 13 | 86.7 |
G | 14 | 93.3 |
LE | 16 | 93.8 |
OF | 9 | 60 |
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1299
ISSN 2229-5518
Discussion
Out of 250 patients, 49 patients blood culture sample were found positive which includes Pseudomonas (22.4%), Staphylococcus (40.8%), Klebsiella (12.2%), Acinetobacter (2.04%), E.coli (16.3%), Proteus mirabilis (2.04%), Candida (2.04%), Salmonella typhi (2.04%) and 201 blood culture sample were negative. In adult, the total positive samples were 72 in which 14 were found to be positive (19.5%). In children, the total samples were 112 in which 20 were found to be positive (17.9%). While in new born individuals there are 66 sample in which 15 were found to be positive (22.7%).
As compared to all these individuals, the populations of children with positive blood culture sample were found to be more i.e 20 as compared to adult (14), and new born (15). The principal hypothesis examined in this study was that the rate of contamination of blood cultures taken at central line would be lower than that of blood cultures taken from peripheral line. Out of the 250 patients, 12.5% were taken during central line insertion, 20.5% were
taken from peripheral veinpuncture.
The process of obtaining blood cultures can be divided broadly into three parts- preparing the phlebotomy site, drawing the blood and inoculating the blood into the culture bottles. All three parts can be sources of blood culture contaminations. However, in the process of drawing blood between central and peripheral blood culture. Central line insertion involves the use of a larger needle which involves considerable manipulations of the skin and subcutaneous tissues during catheter insertion and takes longer than veinpuncture. Approximately, 20% of skin bacteria live within the deeper layers of the dermis and subcutaneous tissue into which topical antiseptics cannot penetrate (Brown, E; R.P. Wenzel & J.O Hendley 1989, Selwyn, S; & H. Ellis 1972) and these bacteria could be contaminating centrally drawn cultures. Similarly, discarding the initial blood volume at phlebotomy for blood cultures decreased blood culture contamination (Patton, R.G; & T. Schmitt 2010). The longer time required for central line insertion also increase exposure to environmental contaminants.
Cultures obtained by peripheral veinpuncture are often drawn for
investigations of fever or leukocytosis,
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1300
ISSN 2229-5518
possibly associated with a lower pretest probability of bacteremia.
Staphylococcus aureus was the most predominant isolates (65%) followed by E.coli (29.3%). This observation is in accordance with reports from two other developing countries (Weber M W, Carlin JB, Gatchalian S, Lehman D, Muhe L, Mulholland EK, WHO young infants study group 2003). Similarily in a study conducted by Martin M Meremikwv et al in Nigeria found that Staphylococcus aureus was isolated in 48.7% and E.coli in 23.4% as the most frequent isolates (Martin M Merimikwv, Chukwuemoka E Nwachukwv Anne E Asuque, Joseph U Okebe 1 and Simon J Utsala 2005). In our study, Staphylococcus aureus was found to be
40.8% followed by pseudomonas 22.4%, while Acinetobacter, Proteus mirabilis, Candida and Salmonella typhi were found to be less i.e 2.04%
Aziz et al reported that staphylococcus aureus (25%) and E.coli (12.1%) as the most pathogenic bacteria recovered from blood sample. While in our study, Staphylococcus aureus(40.8%) and pseudomonas (22.4%) was the most pathogenic bacteria. E.coli, Staphylococcus
aureus and Klebsiella pneumonia were
found to be highly prevalent among new born individuals (Martin M Merimikwv, Chukwuemoka E Nwachukwv Anne E Asuque, Joseph U Okebe 1 and Simon J Utsala 2005). Staphylococcus aureus was found to be more sensitive to Chloramphenicol (87.5%), Ofloxacin (76.5%) and Ciprofloxacin (72%).
While in our study it is found to be more sensitive to Vancomycin (90%), Linezoid (85%), Amikacin (90%), Gentamicin (90%) followed by Levofloxacin (95%) while in gram negative bacteria Imipenem, Meropenem was found to be more sensitive. Ghanshyam D Khumhar (V G Ramachandran and Piyush Gupta 2002) in his study done at India also found that E.coli and Klebsiella is sensitive to Amikacin but Staphylococcus aureus was more sensitive to Vancomycin. While in our study, Pseudomonas shows 100% sensitivity against Levofloxacin.
Automated blood culture is designed to do blood culture for both aerobic and anaerobic cultures. Automated blood cultures comprises of Bac T Alert 3D 120 systems which has a capacity of 60 blood culture bottles. It helps in quick identification of bacterial growth in the
bottles. 24 hours continuous scanning is
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1301
ISSN 2229-5518

done which helps to identify the growth of organisms. It’s easy to use and it is time saving. In manual blood culture, blood culture is done manually i.e 1 ml of sample is taken from blood culture bottle with the help of syringe and subculture it on Mac conkey and blood agar plate. Growth is observed after 24 hours. It is then again subculture for 5 days and final results are noted down. It is time consuming and proper handling should be taken while performing.
IJSER © 2013 http://www.ijser.org

International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1302
ISSN 2229-5518
patients.
.
Lactose and non-lactose fermenting bacteria.
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1303
ISSN 2229-5518
Conclusion
Septicemia is the presence of bacteria in the blood (bacteremia) and is often associated with severe disease. Its alternative name is blood poisoning (bacteremia with sepsis).Out of 250 blood culture samples, 49 patients’ samples were found to be positive which include adult, children and new born patients. Children population with positive blood culture were found to be more as compared to adult and new born. Organisms like Staphylococcus aureus, Pseudomonas aeruginosa, candida, Klebsiella, Proteus mirabilis, Acinetobacter, and salmonella typhi were isolated from these blood culture samples. Each isolated were then treated with antibiotic to observe the sensitivity pattern of these organisms. Each isolates were treated with different
Antibiotic to see the effect or sensitivity
Pattern against those organisms. Out of these isolates Staphylococcus were found to be the pathogenic bacteria. It shows higher sensitivity against Vancomycin, and shows low sensitivity against Amikacin and ciprofloxacin. Subculturing of the positive blood culture is done manually or by using automated blood culture system. Automated blood culture shows accurate result and it shows a quick identifications of organisms present in the sample, while in manually it is time consuming and can sometime lead to false result. Hence, person showing thessymptons of fever may show the sign of septicemia which can be treated or decreased down by using antibiotic against that pathogen.
References
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1304
ISSN 2229-5518
1) Anonymous. 1996. National Nosocomial Infections Surveillance (NNIS) data summary from October 1986-April
1996, issued May 1996. A report from the National Nosocomial Infections Surveillance (NNIS) System. Am. J. Infect. Control 24:380–388.
2) Bates, D. W., L. Goldman, and T. H.
Lee. 1991. Contaminant blood cultures and resource utilization. The true consequences of false-positive results. JAMA 265:365–369.
3) Bates, D. W., L. Goldman, and T. H.
Lee. 1991. Contaminant blood cultures and resource utilization. The true consequences of false-positive results.
4) Bryan, C. S. 1989. Clinical implications of positive blood cultures. Clin. Microbiol. Rev. 2:329–353.
5) Segal, G. S., and J. M. Chamberlain.
2000. Resource utilization and contaminated blood cultures in children at risk for occult bacteremia. Arch. Pediatr. Adolesc. Med. 154:469–473.
6) Souvenir, D., D. E. Anderson, Jr., S.
Palpant, H. Mroch, S. Askin, J. Anderson, J. Claridge, J. Eiland, C. Malone, M. W. Garrison, P. Watson and D. M. Campbell. 1998. Blood cultures positive for coagulase-negative
staphylococci: antisepsis,
pseudobacteremia, and therapy of patients. J. Clin. Microbiol. 36:1923–
1926.
7) Schifman, R. B., and A. Pindur. 1993.
The effect of skin disinfection materials on reducing blood culture contamination. Am. J. Clin. Pathol. 99:
536–538.
Dunagan, W. C., R. S. Woodward, G. Medoff, J. L. Gray III, E. Casabar,M. D. Smith, C. A. Lawrenz, and E. Spitznagel. 1989. Antimicrobial misuse in patients with positive blood cultures. Am. J. Med, 87:253–259.
8) Waltzman, M. L., and M. Harper. 2001.
Financial and clinical impact of false- positive blood culture results. Clin. Infect. Dis. 33:296–299.
9) Thuler, L. C., M. Jenicek, J. P. Turgeon, M. Rivard, P. Lebel, and M. H. Lebel.
1997. Impact of a false positive blood culture result on the management
of febrile children. Pediatr. Infect. Dis. J.
16:846–851
10) Everett E.D, Hirschmann IV 1997.
Transient Bacteremia and endocarditis prophylaxis : a review 1997
11) Weinstein, M. P., L. B. Reller, J. R.
Murphy, and K. A. Lichtenstein.
1983.The clinical significance of positive blood cultures: a comprehensive
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1305
ISSN 2229-5518
analysis of 500 episodes of bacteremia and fungemia in adults. I. Laboratory and epidemiologic observations. Rev. Infect. Dis. 5:35–53.
12) Weinstein, M. P. 2003. Blood culture contamination: persisting problems
and partial progress. J. Clin. Microbiol.41:2275–2278.
13) Calfee, D. P., and B. M. Farr. 2002.
Comparison of four antiseptic preparations
for skin in the prevention of contamination of percutaneously drawn
blood cultures: a randomized trial.
14) Norberg, A., N. C. Christopher, M. L.
Ramundo, J. R. Bower, and S. A. Berman. 2003. Contamination rates of blood cultures obtained by dedicated phlebotomy vs intravenous catheter. JAMA 289:726–729.
15) Rubin, L. G., P. J. Sanchez, J. Siegel, G.
Levine, L. Saiman, W. R. Jarvis, et al.
2002. Evaluation and treatment of neonates with suspected late-onset sepsis: a survey of neonatologists’ practices. Pediatrics 110:e42.
16) Galdbart, J. O., A. Morvan, N.
Desplaces, and N. el Solh. 1999. Phenotypic and genomic variation among Staphylococcus
epidermidisstrains infecting joint prostheses. J. Clin. Microbiol. 37:1306–
1312.
17) Van Wijngaerden, E., W. E. Peetermans, S. Van Lierde, and J. Van Eldere. 1997. Polyclonal staphylococcus endocarditis. Clin. Infect. Dis. 25:69–71
18) Sharma, M., K. Riederer, L. B. Johnson, and R. Khatib. 2001. Molecular analysis of coagulase-negative Staphylococcus isolates from blood cultures: prevalence of genotypic variation and polyclonal bacteremia. Clin. Infect. Dis. 33:1317–
1323.
19) Van Wijngaerden, E., W. E.
Peetermans, S. Van Lierde, and J. Van
Eldere.
1997. Polyclonal staphylococcus endocarditis. Clin. Infect. Dis. 25:69–71
20) Mirrett, S., M. P. Weinstein, L. G.
Reimer, M. L. Wilson, and L. B. Reller.
2001. Relevance of the number of positive bottles in determining clinicalsignificance of coagulase- negative staphylococci in blood cultures. J. Clin.Microbiol. 39:3279–3281.
21) Saito, T., K. Senda, S. Takakura, N.
Fujihara, T. Kudo, Y. Iinuma, M. Tanimoto, and S. Ichiyama. 2003. Detection of bacteria and fungi in BacT/
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1306
ISSN 2229-5518
Alert standard blood-culture bottles. J. Infect. Chemother. 9:227–232.
22) Khatib, R., K. M. Riederer, J. A. Clark, S. Khatib, L. E. Briski, and F. M. Wilson. 1995. Coagulase-negative staphylococci in multiple blood cultures: strain relatedness and determinants of same-strain bacteremia. J. Clin. Microbiol. 33:816–820.
23) Catton, J. A., B. M. Dobbins, P. Kite, J.
M. Wood, K. Eastwood, S. Sugden, J. A. Sandoe, D. Burke, M. J. McMahon, and M. H. Wilcox. 2005. In situdiagnosis of intravascular catheter-related bloodstream infection: a comparisonof quantitative culture, differential time to positivity, and endoluminalbrushing. Crit. Care Med. 33:787–791.
24) Franklin, J. A., A. H. Gaur, J. L. Shenep, X. J. Hu, and P. M. Flynn. 2004. In situ diagnosis of central venous catheter- related bloodstream infection without peripheral blood culture. Pediatr. Infect. Dis. J. 23:614–618.
25) Innes, G., K. Roland, E. Grafstein, and J.
M. Christenson. 2000. Utility of blood cultures in the emergency department. Acad. Emerg. Med. 7:579
26) Evans, M. R., A. L. Truant, J. Kostman, and L. Locke. 1991. The detection of
positive blood cultures by the BACTEC
NR660. The clinical importance of four- day versus seven-day testing.Diagn. Microbiol. Infect. Dis. 14:107– 110.
27) Huang, A. H., J. J. Yan, and J. J. Wu.
1998. Comparison of five days versus seven days of incubation for detection of positive blood cultures by the Bactec
9240 system. Eur. J. Clin. Microbiol. Infect. Dis. 17:637–641.
28) Kurlat, I., B. J. Stoll, and J. E.
McGowan, Jr. 1989. Time to positivity for detection of bacteremia in neonates. J. Clin. Microbiol. 27:1068–1071.
29) Mozes, B., D. Milatiner, C. Block, Z.
Blumstein, and H. Halkin. 1993. Inconsistency of a model aimed at predicting bacteremia in hospitalized patients. J. Clin. Epidemiol. 46:1035–
1040
30) Haimi-Cohen, Y., S. Shafinoori, V.
Tucci, and L. G. Rubin. 2003. Use of incubation time to detection in BACTEC
9240 to distinguish coagulase negative staphylococcal contamination from infection in pediatric blood cultures. Pediatr. Infect. Dis. J. 22:968–974.
31) Bates, D. W., K. E. Pruess, and T. H.
Lee. 1995. How bad are bacteremia and sepsis? Outcomes in a cohort with suspected bacteremia. Arch. Intern. Med.
155:593–598.
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 11, November-2013 1307
ISSN 2229-5518
32) Alpern, E. R., E. A. Alessandrini, L. M.
Bell, K. N. Shaw, and K. L. McGowan.
2000. Occult bacteremia from a pediatric emergency department current prevalence, time to detection, and outcome. Pediatrics 106:505–511.
33) Bandyopadhyay, S., J. Bergholte, C. D.
Blackwell, J. R. Friedlander, and H. Hennes. 2002. Risk of serious bacterial infection in children with fever without a source in the post- Haemophilusinfluenzae era when antibiotics are reserved for culture- proven bacteremia. Arch. Pediatr. Adolesc. Med.156:512–517. (Erratum,
156:749.)
34) Baraff, L. J., J. W. Bass, G. R. Fleisher, J. O. Klein, G. H. McCracken, Jr., K. R. Powell, D. L. Schriger, et al. 1993. Practice guideline for the managementof infants and children 0 to 36 months of age with fever without source. Ann. Emerg. Med. 22:1198–1210. (Erratum,
22:1490.)
35) Ammann, R. A., A. Hirt, A. R. Luthy, and C. Aebi. 2003. Identification of children presenting with fever in
chemotherapy-induced neutropenia at low risk for severe bacterial infection. Med. Pediatr. Oncol. 41:436–443.
36) Johnson, J.E; and J.A Washington III
1976. Comparison of direct and standardized antimicrobial susceptibility testing of positive blood cultures.
37) Kiehn, T.E; C. Capitolo and D.
Armstrong 1982. Comparison of direct and standard microtiter broth dilution susceptibility testing of blood culture isolates.
38) Mirrett, S; and L. BarathReller 1979.
Comparison of direct and standard antimicrobial disk susceptibility testing for bacteria from blood.
39) Wegner, D.L; C.R Mathis and T.R Neblett 1979. Direct method to determine the antibiotic susceptibility of rapidly growing blood pathogens.
40) Reller. L.B; P.R Murray and J.D Maclowry 1982. Cumitech 1A,, Blood cultures II. Coordinating ed.. J.A Washington II. American society for microbiology.
IJSER © 2013 http://www.ijser.org