The research paper published by IJSER journal is about A Simple DNA Extraction Method For PCR Amplification From Young Needles Of Cedrus Deodara 1
ISSN 2229-5518
A Simple DNA Extraction Method For PCR Amplification From Young Needles Of Cedrus Deodara
Kamraj Singh*, Shalini singh, Rahul Sahu
ABSTRACT- Molecular based genetic analysis requires good quality of DNA in sufficient quantity to generate robust DNA, fingerprinting. A simple and reliable DNA extraction method for young needles (Leaves) of C. deodara has been developed in our laboratory. The NACl, and PVP were used to remove polysaccharides and polyphenols during DNA purification. The oil and proteins of young needles were removed only through centrifugation in this method. The good result of SSR (Simple sequence repeat) molecular markers on the DNA of young needles of another C. deodara indicating the DNA extracted from Young needles was free from common contaminating compounds. In conclusion this method could be widely used in DNA extraction from young needles of C. deodara.
KEYWORDS- DNA, Extraction, Cedrus deodara, SSR, Liquid nitrogen, β-mercaptoethanol
—————————— ——————————
1. INTRODUCTION
edrus deodara is an ecologically and economically important
forest tree species of western Himalayas and is considered on the endangered conifer species in this region micro-satellite markers (SSR) were used to study the genetic variation within C. deodara. Recently, this species has been used for molecular analysis, such as genetic variation analysis (Seysis et al., 2003, Pallet et al., 2006) and gene mapping (Lombard and deloureme,

2001, kole et al., 2002, Javinfar et al. 2006). Most of the plant DNA extraction methods are essentially either a CTAB method (Murray and Thompson.1980) or an SDS- potassium acetate method (Delloporta et al., 1983) However, these methods are not appropriate for a large number of DNA extractions in a short period of time such as for a large number such as for marker- assisted selection in rapeseed breeding for their time consuming, various simple method by using the leaves as materials have been developed such as a direct amplification of needles tissue (Berthomilu and Mayer, 1991) , boiling method (Thomson and Henry, 1995, Ikeda et al., 2001) and alkali treatment method (Xin et al., 2003). In other Species (Brassica napus) we have successfully tested several procedures for DNA extraction by using dry seeds as materials (Horn, 1992, Li et al., 1994) but few methods were developed for young needles C. deodara. In this paper, we reported a simple DNA extraction for young needles of C. deodara; it could be finished very quickly and could produce relatively high quality DNA for SSR molecular markers.
Kamraj singh tomar*, MSc. Biotechnology, Dept. Of Biotechnology,Dolphin (PG) Instt. Of Biomedical & Natural Sciences, Dehradun (Uttarakhand)#, kamrajmbt86@gmail.com
Shalini singh, Dept. Of Biotechnology #.
Rahul sahu, MSc. Biochemistry, Dept. Of Biochemistry#
rahul_sahu1309@yahoo.co.in
R © 2012
www.ijser.org
2. MATERIALS AND METHODS
2.1 Materials: Young needles of C. deodara were used for
DNA extraction
2.2 Methods:
2.2.1 DNA extraction method for young needles
(Leaves)
Prepared extraction buffer (100mM Tris HCL (pH 8.0),
20 mM EDTA, 5mM Ascorbic acid, 2% C-TAB, 2% PVP).
To 1ml of preheated extraction buffer, 3µl of β- mercaptoethanol in 2 ml eppendorf tubes was added.
The leaves were ground in alcohol wiped mortar and pestle with continuous addition of liquid nitrogen and the powder were transferred to the eppendorf tubes containing pre-heated extraction buffer.
Vortex the eppendorf tubes for 15 seconds.
Incubated in water bath for 30 minutes at 600C.
Added 500µl solution of chloroform : isoamyl alcohol
(24:1) followed by inversion.
Centrifuged at 13,000 rpm for 5 min (40C).
Supernatant was transferred to 1.5 ml eppendorf tubes carefully.
Added 500µl of cold (-200C) isopropanol and mixed by inversion.
Incubated overnight at - 200C. Then centrifuged at 40C at
13,000 rpm for 15 minutes to obtain pellet.
Added 998µl of 76% ethanol and 2µl of ammonium acetate followed by its inversion for minutes on gel rocker.
Again centrifuged for 5 minutes and added 500µl of 70% ethanol and centrifuge for 15 minutes at 40C at 13,000 rpm.
Vacuum dried the pellet and dissolved in 100-200µl of
TE buffer.
The research paper published by IJSER journal is about A Simple DNA Extraction Method For PCR Amplification From Young Needles Of Cedrus Deodara 2
ISSN 2229-5518
2.2.2SSR analysis: -
5SSR primers, Pt15169, Pt26081, Pt3020, Pt71936 and Pcp
26106 were used for SSR analysis. The PCR procedure was followed the method of Saal et al. (2001) and LI et al. (2005). The PCR reaction profiles were as followed: 940C at 60 sec followed by 35 cycles of 60 sec at 940C, 60 s at 610C and 1.5 min at 720C, extension at 720Cfor 10 min. and then head at
40C. The 20µl formamide loading Buffer was added into
selective amplification reaction product, the mixed sample were denatured in 950C for 5 min. cooled on ice and then analysis in 6% PAGE gel. The gel was stained with silver staining kits according to the manufacturer’s instructions.
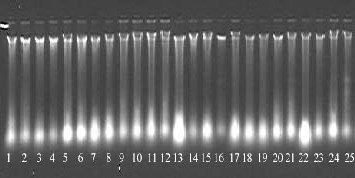
Fig1: Gel image of total genomic DNA isolated from young needles of C. Deodara using modified protocol. Lance 1-25 showing isolated Genomic DNA.
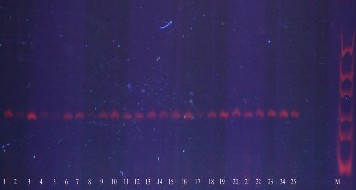
Fig2: SSR pattern obtained from DNA extracted from modified protocol. 25 different DNA samples (Lanes 1-25) amplified using prime Pt 26081. Lane-M molecular weight marker.
3. RESULTS AND DISCUSSION:-
The materials that used in DNA extraction were fresh needles in most cases (Murray and Thompson, 1980; Dellaporta et al.,
1983; Saghai- Maroof et al., 1984; Doyle and Doyle, 1987; Lange et al., 1998). The DNA extraction method by using the seeds as material was also reported in some crops, such as in
rice (Chunwonges et al., 1993; Peng et al., 2002), corn
(McDonald et al., 1994) and Soyabean (Young et al., 2003). We have obtained higher quality of DNA from fresh needles of C. deodara by using the protocol outline above. The fatty acid and proteins were the main components of the needles of C. deodara. For fatty acid has lower density and non- polar characteristic, it could easily be distinguished from the aqueous phase when it involved into centrifugation. The phenol and chloroform were frequently used for protein removing in custom DNA extraction method; it needed only need a short centrifugation to separate DNA from all the other contaminants in our present protocol, for the most of proteins removed in the insoluble precipitate. The 1M Nacl was added into extraction buffer to remove the polysaccharides by increasing their solubility in ethanol (Fange et al., 1992). In order to remove polyphenols from the fresh needles, the PVP was added to the extraction buffer according to the result of Maliyakal (1992).
By using the DNA extraction protocol outlined above, we have obtained higher quality of DNA from fresh needles of C. deodara with A260/ A280 between 1.7 and 2.0, which indicated that protein could be removed through centrifugation. The DNA was further analysed in agarose gel, there was no DNA degradation and the average size of the bands was about 20 kb (Fig.1). The DNA extracted from fresh needles was successfully used in SSR analysis. Good amplification result of SSR molecular markers also verified the good quality of these 25 DNA samples (Fig. 2) RNA could be removed by digesting the sample with RNAse (Horn.
1992). In our experiment, we found that the RNA did not interfered with marker development, so the step for add the RNAse A may be removed in our developed protocol. In conclusion, our newly developed DNA extraction protocol can produce clean and high –quality DNA that is suitable for SSR molecular analysis in C. deodara.
ACKNOWLEDGMENT
We want to express our sincere thanks to Director, Dolphin (PG) Instt. Of Biomedical & Natural Sciences, Dehradun (Uttarakhand)
REFERENCES
1. Berthomieu P. and C. Meyer, 1991. Direct amplification of plant genomic DNA from leaf and root Pieces using PCR. Plant Mol. Biol., 17: 555-557.
2. Chunwonges J., G.B. Martin and S.D. Tanksley,
1993. Pre-germination genotype screeing using PCR
amplification of half –seeds. Thero. Applied Genet.,
86: 694-698.
IJSER © 2012
http://www.ijser.org
The research paper published by IJSER journal is about A Simple DNA Extraction Method For PCR Amplification From Young Needles Of Cedrus Deodara 3
ISSN 2229-5518
3. Dellaporta S., J. Wood and J.B Hicks, 1983. A plant DNA minipreparation: Version II. Plant Mol. Biol. Rept., 1: 19-21.
4. Doyle J.J and J.L. Doyle, 1987. Arapid DNA isolation procedure for small quantities of fresh leaves tissue. Phytochem. Bull., 19: 11-15 .
5. Fung G., S. Hammar and R. Rebecca, 1992. A quick
and in expensive method for removing polysaccharides from plant genomic DNA. Biotechnology, 13:52-56.
6. Horn P. and A. Rafalski, 1992. Non-destructive RAPD genetic diagnostics of microspore-derived Brassica embryos. Plant Mol. Biol. Rep., 10: 285-
293.
7. Ikeda N., N.S. Bautista, T. Yamada, O. Kamijima and T. Ishii, 2001. Ultrasimple DNA extraction method for marker-assisted selection using microsatellite markers in rice. Plant Mol. Biol. Kept.,
19: 27-32. Javidfar, F., V.L.
8. Kole C., C.E. Thormann, B.H. Karlsson, J.P. Palta, P.
Gaffney, B. Yandell and T.C. Osborn, 2002. Comparative mapping of loci controlling winter survival and related traits in oilseed Brassica rapa and B. napus. Mol. Breed., 9: 201-210.
9. Lange D.A., S. Penuela and R.L. Denny, 1998. A plant DNA isolation protocol suitable for polymerase chain reaction based marker-assisted breeding. Crop. Sci., 38: 217-220.
10. Li J., B.Z. Shen and J.X. Han, 1994. An effective way of total DNA extraction from Rapeseed. J.
Huazhong. Agric. Univ., 13: 521-523.
11. Li M., C. Zhang, Z. Li and J. Meng, 2005. The analysis of the biological characters in hexaploid hybrids Derived from Brassica carinata and B. rapa. Acta. Agronomica Sinica, 12: 1579-1585.
12. Li M., Z. Li, C. Zhang, W. Qian and J. Meng, 2005.
Reproduction and cytogenetic characterization of interspecific hybrids derived from crosses between Brassica carinata and B. rapa. Theor. Applied Genet.
110: 1284-1289.
13. Lombard V. and R. Delourme, 2001. A consensus
linkage map for rapeseed (Brassica napus L.): Construction and integration of three individual maps from DH populations. Theor. Applied Genet., 103:
491-507.
14. Maliyakal E.J., 1992. An efficient method for isolation of RNA and DNA from plants containing polyphenolics. Nucl. Acids Res., 20: 2381-2381.
15. McDonald M.B., L.J. Elliot and P.M. Sweenery,
1994. DNA extraction from dry seeds for RAPD analyses in varietal identification studies. Seed Sci. Technol., 22: 171-176.
16. Murray M.G. and W.F. Thompson, 1980. Rapid isolation of high molecular weight DNA. Nucleic Acids Res., 8: 4321-4325.
17. Peng S., Q. Yan, X. Wang and C. Zhang, 2002. DNA extraction from single seeds and optimizing RAPD procedure in rice. J. Shanghai. Jiaotong. Univ. (Agricultural Science), 20: 34-37.
18. Ripley V. Roslinsky, H. Zeinali and C. Abdmishani,
2006. Identification of molecular markers associated with oleic and linolenic acid in spring oilseed rape (Brassica napus). Plant Breed., 125: 65-71.
19. Saal B., J. Plieske, J. Hu, C.F. Quiros and D. Stress,
2001. Micosatellite markers for genome analysis in Brassica. II. Assignment of rapeseed micosatellites to the A and C genomes and genetic mapping in Brassica oleracea L. Theor. Applied Genet., 102:
695-699.
20. Saghai-Maroof, M.A., K.M. Soliman, R.A. Jorgensen and R.W. Allard, 1984. Ribosomal DNA spacer- length polymorphisms in barley: Mendelian inheritance, chromosomal location and population dynamics. Proc. Malt. Acad. Sci. USA., 81: 8014-
8018.
21. Seyis F., R.J. Snowdon, W. Luhs and W. Friedt,
2003. Molecular characterization of novel resynthesized rapeseed lines and analysis of their genetic diversity in comparison with spring rapeseed cultivars. Plant Breed., 122: 473-478.
22. Thomson D. and R. Henry, 1995. Single-step protocol for preparation of plant tissue for analysis by PCR. Biotechnology, 19: 394-400.
23. Xin Z., J.P. Veiten, M.J. Oliver and J.J. Burke, 2003.
High-throughput DNA extraction method suitable for
PCR. Biotechnology, 34: 820-825.
24. Yang S., L. Zhang and H. Duan, 2003. Rapid extraction of soybean for PCR from dry seed. Soybean Sci., 22: 151-153.
IJSER © 2012
http://www.ijser.org
International Journal of Scientific & Engineering Research Vo lume 3, Issue 12, December-2012 4
ISSN 2229-5518
IJSER lb)2012
httr: /Jwww.i jser.
org


